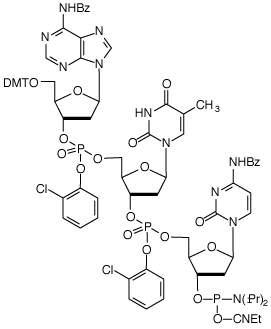
Trimer Phosphoramidites – Tools for fine-tuning protein function Introducing mutations in existing proteins can be used to fine-tune almost any desired property, such as improved stability to high temperatures, denaturants, or non-aqueous solvents; higher affinity binding to a target molecule; increased rates of enzymatic reactions; or changes of specificities. However, generating and finding these improved proteins can be a difficult task. Now for the first time, new tools are available to radically improve the efficiency of this process – Trimer Phosphoramidites.1-4 Covering all 20 amino acids, Trimer Phosphoramidites (Figure 1) allow mutation of a gene at the codon level rather than at individual bases. Therefore, unlike other methods of mutagenesis, Trimer Phosphoramidites lead to no codon bias, no frame-shift mutations, and no production of stop codons, making them highly efficient tools for the exploration of sequence space in protein regions that are important for function. Trimer phosphoramidites can be added during synthesis using standard DNA synthesis chemistry. A Reaction Factor (RF) has been determined for each Trimer to compensate for differences in their relative rate of reaction during coupling. It is therefore possible to introduce an equimolar mix of all 20 amino acid codons, or subsets thereof, at any location within the sequence.
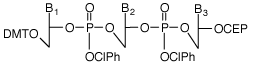
Of course, there are cheaper alternatives for introducing mutations, but they are far less effective and may cause considerably more expense in downstream screening for desired mutations. One of the most popular choices is to make pools of degenerate oligonucleotides, which can be incorporated into the genes as cassettes or by PCR by using the degenerate oligo as a primer.5 Degenerate oligonucleotides are synthesized as a mixture of A/C/G/T phosphoramidites (N) at the site of the codons to be mutated. Problems arise, though, from using an equimolar solution of each base. First there is a coding bias. Out of the 64 possible codon combinations of A, C, G and T, 18 code for leucine, arginine or serine, but only 2 for tryptophan or methionine. As a result, only 3% of the mutagenic oligonucleotides will contain methionine or tryptophan, and over 28% will contain either leucine, arginine or serine. In addition, the three nonsense codons will lead to chain termination in 4.7% of the sequences. There are ways to help this situation. For instance, using two degenerate mixes of bases, N and G/C, on the DNA synthesizer to insert NNG/C into the sequence will halve the number of the most degenerate codons, but still code for all 20 amino acids. However, still 59% of the clones will code for just eight amino acids and 3% will have a stop codon inserted. The generation of redundant sequences and stop codons makes searching a clonal library inefficient.
Trimer phosphoramidites offer an elegant solution6 that circumvents these problems of codon bias, frame-shift mutations and stop-codon production – even in nonsaturating conditions.7
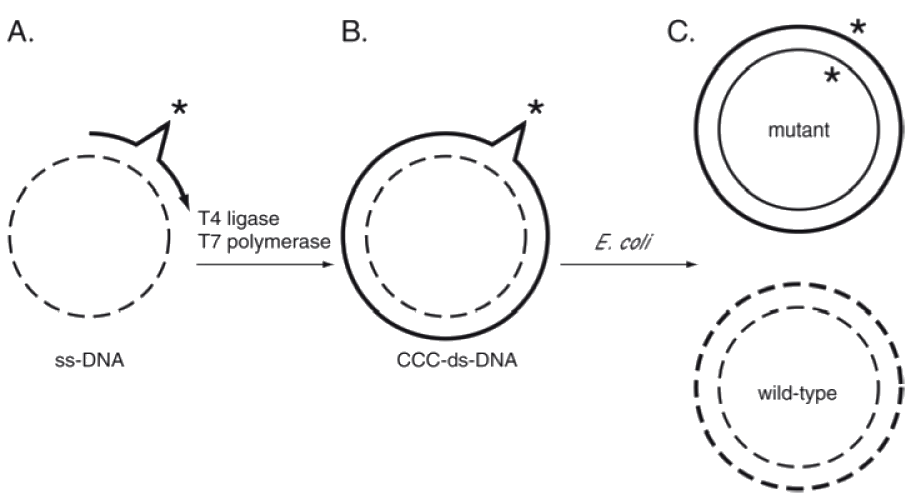
Once a mutagenic oligonucleotide has been synthesized using Trimer Phosphoramidites, there are a variety of means of introducing it into a plasmid for construction of a library. A very efficient means of mutagenesis has been reported8 as shown in Figure 2. A synthetic oligonucleotide (solid line) is annealed to a circularized dU-containing ssDNA template (dashed line) that was obtained from dut -/ung - E. coli strain that lacks Uracil DNA glycosylase. The mismatched variable region is flanked by perfectly complementary sequences (A). Covalently closed circular DNA (CCC-ds DNA) is obtained by the action of the T7 polymerase and T4 ligase (B). When the CCC-ds-DNA is introduced into ung+ E. coli, the template DNA, containing dU, is preferentially destroyed, leading to the enrichment of the mutant plasmid (C).
We thank Sachdev Sidhu, Ron Godiska and Paul Gaytan for reviewing this document and for their many helpful suggestions.