Glen Report 18.17: Potent Inhibition of Cytosine-5-methyltransferases
The methylation of cytosine in the CpG motif of DNA almost invariably leads to a reduction in gene expression and is one of the most prevalent methods for the epigenetic regulation of genes. Cytosine-5-methyltransferases are found in everything from archaebacteria to mammals and when the regulation of cytosine-5-methyltransferases goes awry, cancer can result. DNA methyltransferases were found to be essential for cancer cell survival1 and to work in concert with p532. The mechanism of action for this family of enzymes involves attack of a cysteine thiol group on the C6 position of cytosine, leading to a transient dihydrocytosine intermediate, which then facilitates the nucleophilic attack by C5 on the activated methyl group of the S-adenosyl-L-methionine cofactor.
As with many enzymes, the intermediate can be trapped using a suicide substrate and 5-fluoro-cytosine3 has been used extensively in this role. An alternate strategy is to use a transition-state mimic that binds to the active site with high affinity. An excellent candidate was found in 5-aza-5,6-dihydrocytosine. Despite not being covalently bound to the enzyme, it was found4,5 to be a more potent inhibitor of cytosine-5-methyltransferases than 5-fluoro-cytosine. Happily, 5-aza-5,6-dihydro-dC is amenable to conversion to the phosphoramidite6 and is compatible with standard oligonucleotide synthesis and deprotection conditions. We enthusiastically offer this monomer (1) as a new tool for use in methyltransferase research. It would be remiss of us if we did not specifically highlight the work done on the chemistry and biology of 5-azacytosines and their interaction with cytosine-5-methyltransferase over many years by Victor Marquez and co-workers.
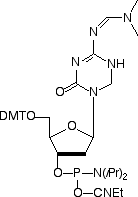
References:
- N. Beaulieu, et al., J Biol Chem, 2002, 277, 28176-81.
- P.O. Esteve, H.G. Chin, and S. Pradhan, Proc Natl Acad Sci U S A, 2005, 102, 1000-5.
- A.M. MacMillan, L. Chen, and G.L. Verdine, J. Org. Chem., 1992, 57, 2989-2991.
- G. Sheikhnejad, et al., J Mol Biol, 1999, 285, 2021-2034.
- V.E. Marquez, et al., Antisense Nucleic Acid Drug D, 1999, 9, 415-421.
- A.J. Goddard and V.E. Marquez, Tetrahedron Letters, 1988, 29, 1767-1770.
5-aza-5,6-dihydro-dC-CE Phosphoramidite (10-1511) has been discontinued
Amino-Modifier C6 dG
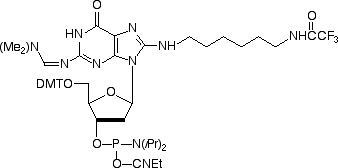
When we introduced Amino-Modifier C6 dA in 2003, we were aware that the structure was not optimal and that there would be some destabilization of the base pair with dT. Indeed, the base pair was destabilized by 2 °C per insertion but binding was still found to be specific for dA. We noted at the time that the optimal attachment point in purines was probably the 7-position of the 7-deaza purine and preferably using the 7-deaza-8-aza analogue. However, these analogues are very difficult to make and are consequently very expensive. In the case of Amino-Modifier C6 dA, we have been encouraged by its popularity despite these shortcomings. So we felt it would be reasonable to offer the dG analogue, Amino-Modifier C6 dG. This dG analogue was found to maintain specificity for dC in oligos but the melting temperature was reduced by 3 °C per insertion relative to the regular dG-dC base pair. (Melting temperatures for oligos containing a mismatch with dA, dG or T opposite dG were reduced by an additional 6-7°C.) Again, these structural characteristics may not be the ideal but Amino-Modifier C6 dG maintains a fair balance of performance and price.
Amino-Modifier C6 dG (10-1099) has been discountinued
- Glen Report 18.11: AP-dC - a Cytosine Analogue Capable of Clamp-like Binding to Guanine - G-clamp
- Glen Report 18.12: Trimer Phosphoramidites – Tools for fine-tuning protein function
- Glen Report 18.13: Sulfurizing Reagent II - Stable in Solution and Optimized for RNA Sulfurization
- Glen Report 18.14: Caps for Increased Duplex Stability and Base-Pairing Fidelity at Termini
- Glen Report 18.15: Synthesis of Branched DNA with a Comb Structure
- Glen Report 18.16: Fluorous Affinity Purification of Oligonucleotides
- Glen Report 18.17: Potent Inhibition of Cytosine-5-methyltransferases
- Glen Report 18-1 Supplement: Sulfurizing Reagent II and its use in Synthesizing Oligonucleotide Phosphorothioates

