Glen Report 32.15: Application Note — Trimer Phosphoramidites
Glen Research offers a unique line of products known as trimer phosphoramidites. These are trinucleotide reagents that allow customers to effectively synthesize oligonucleotides based on amino acid codons rather than individual nucleotides.1 For researchers looking to generate oligonucleotide libraries for mutagenesis, these reagents avoid stop codons and amino acid redundancy. In theory, such libraries can be obtained via a split and pool synthesis workflow without trimer phosphoramidites, but this is very awkward to perform in practice for a large number of codons or longer codon-containing regions.
Glen Research offers a total of 29 trimer phosphoramidites that cover all 20 standard amino acid codons in the sense and antisense directions and are optimized for E. coli expression. We also offer two standard trimer phosphoramidite mixes, both for sense trimers: Mix 2, 13-1992-xx, which contains an equal mixture of 19 codons (no cys), and Mix 1, 13-1991-xx, which contains an equal mixture of 20 codons. For other ratios, we can custom prepare ready-to-use reagents as desired. Alternatively, customers may purchase the trimers individually and then construct their own mixes. Those who do decide to pursue this latter option will need to be mindful of the different coupling rates that the various trimers exhibit, as detailed in Glen Report articles 16.25 and 25.12.
Over the years, our customers have published some exciting results using libraries synthesized with our trimer phosphoramidite mixes. The Regan Group at Yale developed a new screening method for protein affinity agents.2 The technique is based on splitting green fluorescent protein (GFP) into 2 halves and fusing a peptide library and a target peptide to the GFP fragments (Figure 1). During screening, if a member of a library binds to the desired peptide, then the two halves of GFP are brought together resulting in detectable fluorescence. The researchers constructed a peptide library based on the tetratricopeptide repeat (TPR) as a framework (34 amino acids, aa). In the library, seven positions were randomized with our Trimer Phosphoramidite Mix 2 (no cys). This library was fused with the C-terminal fragment of GFP (CGFP) and screened against several targets in E. coli: c-Myc epitope tag (10 aa), full length Dss1 (70 aa), and the C-terminal epitope of Dss1 (19 aa), each of which was fused to the N-terminal fragment of GFP (NGFP). Screening was performed in E. coli, and bacteria that were fluorescent were identified and isolated by fluorescence activated cell sorting (FACS). For each of the targets, two rounds of iterative selection successfully isolated binders that bound specifically with Kds in the micromolar range. 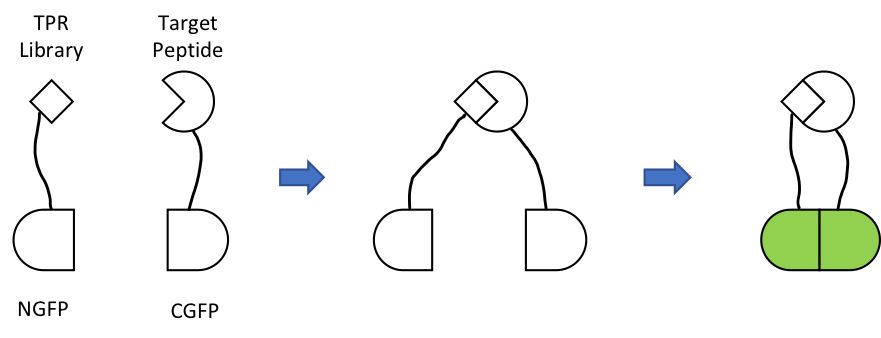
Figure 1. Split-GFP reassembly assay
In another investigation, the Sidhu Group at the University of Toronto examined the importance of certain regions of antibodies in terms of antigen recognition.3 Antibodies are typically “Y-shaped” and consist of 2 heavy and 2 light chains held together by disulfide bonds (Figure 2). At the top of the Y are 2 identical antigen binding domains, and each of these domains contains 6 complementarity-determining regions (CDRs), 3 from the light chain and 3 from the heavy chain. In nature, the third CDR of the heavy chain (CDR-H3) has been shown to be the most important, although it is unclear why. Notably, the location of the third CDR of the light chain (CDR-L3) is positioned to potentially play as big of a role as CDR-H3, meaning that genetics may be the underlying reason for the importance of CDR-H3. The researchers assembled antigen-binding libraries that contained diversity generated with a Custom Trimer Phosphoramidite Mix of Tyr/Ser/Gly/Ala/Phe/Trp/His/Val/Pro 5:4:4:2:1:1:1:1:1. Using this library and phage display, the researchers were able to generate many functional antibodies for a range of different antigens. These synthetic antibodies were analyzed in terms of antigen binding and sequence content, and the results were the opposite of what is observed in nature. CDR-L3 was shown to be more important than CDR-H3. To follow up on these experiments, shotgun alanine-scanning and X-ray crystallography were used to further characterize the CDR to antigen interactions.
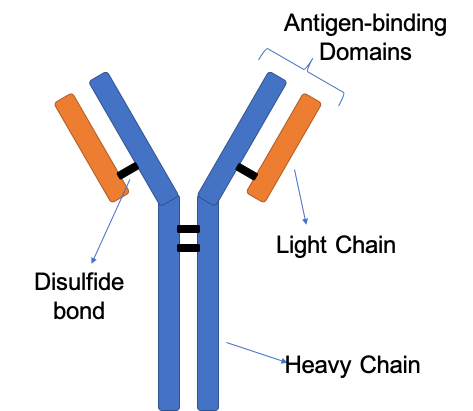
Figure 2. Antibody structure
In a third publication, the Ellington Group at the University of Texas at Austin described a directed evolution method called compartmentalized partnered replication.4 This technique couples the expression of a protein of interest to the expression of Taq DNA polymerase (Figure 3). The authors chose T7 RNA polymerase (T7 RNAP) as a proof of principle. In this setup, Taq DNA polymerase expression would be dependent on T7 RNAP binding onto the promotor. The investigation began with the generation of a plasmid library of T7 RNA polymerase variants, where six of the amino acids in the specificity loop were randomized with oligonucleotides constructed from Trimer Phosphoramidite Mix 1. This library, along with Taq DNA polymerase plasmids, was transformed into E. coli. The bacteria were then transferred to a water-in-oil emulsion in which a large number of water droplets or “compartments” were present. Each droplet statistically contained a single cell as well as PCR primers. Only the cells with desirable mutants would drive expression of Taq DNA polymerase. To measure the amount of Taq DNA polymerase produced, PCR was carried out on the emulsions. Heating during PCR lysed the cells, and the primers that were in the droplet directed the amplification of the T7 RNAP. Each droplet was effectively a single screening experiment. After PCR, the emulsion was collapsed, and the PCR products were isolated. The resulting mutant pool was then used to construct an enriched plasmid pool for another round of compartmentalized partnered replication. After 4 rounds of this, the sequence pool was very similar to the native T7 RNAP sequence.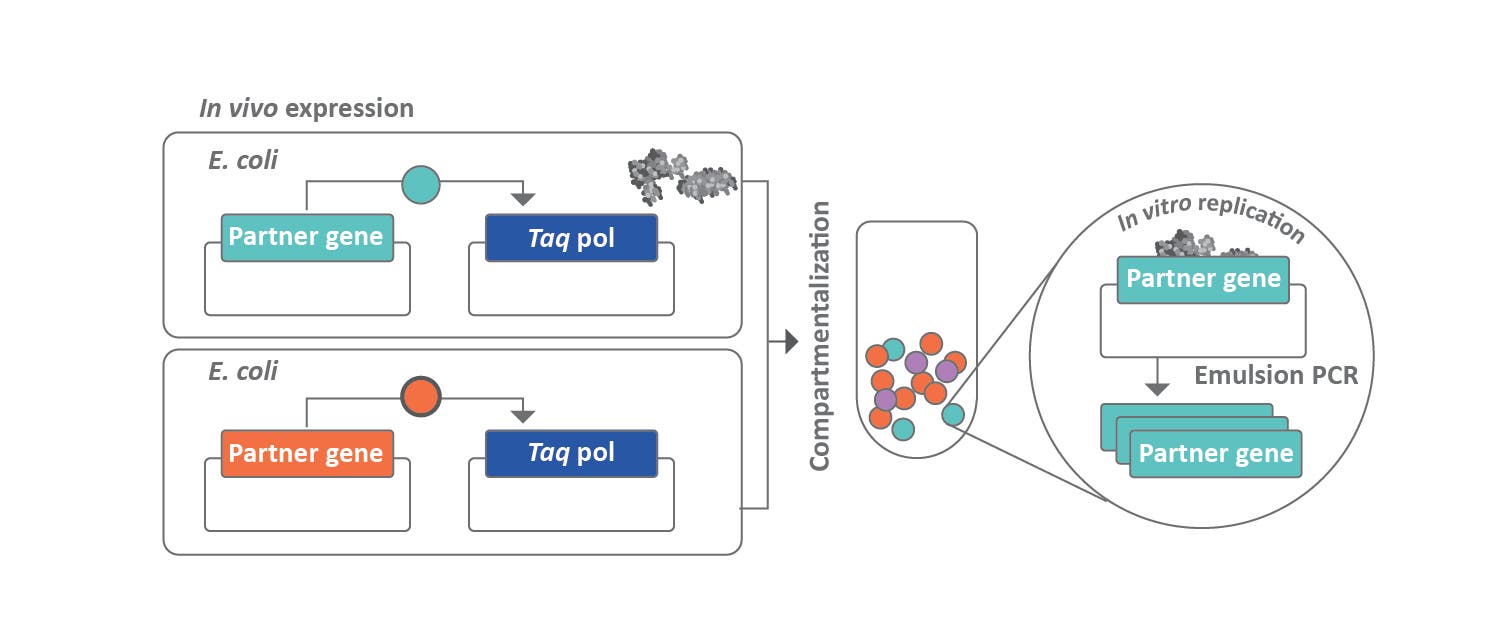
Figure 3. General compartmentalized paired replication concept. Partner genes are shown as either active (green) or inactive (red).
As a follow up experiment, the aforementioned original mutant library was paired with a mutated promotor for Taq DNA polymerase. This required 16 more rounds of selection, several of which involved the use of error-prone PCR to derive mutants that were not available in the original library. In the end, a mutant T7 RNAP/mutant Taq DNA polymerase promotor pair that gave comparable expression levels to the respective native/native pair was characterized.
In each of these investigations, the use of trimer phosphoramidite mixes allowed researchers to create defined sequence libraries that maximized the efficiency of the screening methods they used. No sequence space was lost due to undesired amino acids or stop codons, and the ratios of the amino acids were fully customizable. For those who are still using degenerate oligonucleotides for mutagenesis library construction, it might be a good time to consider using trimer phosphoramidites.
References
- A. Yagodkin, et al., Nucleosides, Nucleotides and Nucleic Acids, 2007, 26, 473 - 497.
- M.E. Jackrel, A.L. Cortajarena, T.Y. Liu, and L. Regan, ACS Chem Biol, 2010, 5, 553-62.
- H. Persson, et al., J Mol Biol, 2013, 425, 803-11.
- J.W. Ellefson, et al., Nat Biotechnol, 2014, 32, 97-101.
Product Information
- Glen Report 32.11: Thiazole Orange as a Fluorogenic Reporter in Oligonucleotide Probes
- Glen Report 32.12: New Product — Thiazole Orange NHS Ester
- Glen Report 32.13: New Product — 2’-Fluoro-Inosine-CE Phosphoramidites
- Glen Report 32.14: New Product — 2'-MOE RNA Phosphoramidites
- Glen Report 32.15: Application Note — Trimer Phosphoramidites
- Glen Report 32.16: Technical Snippets

