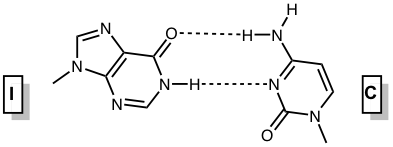
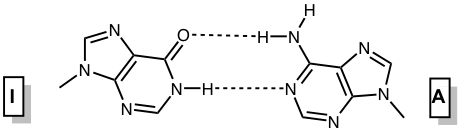
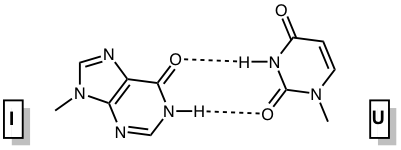
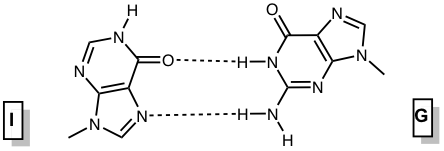
Inosine and 2’-deoxyinosine are non-standard, naturally occurring nucleotides that contain the nucleobase hypoxanthine. Inosine is found in tRNA wobble positions and processed mRNA, while 2’-deoxyinosine is a result of undesired 2’-deoxyadenosine deamination in DNA that needs to be repaired. Inosine is considered a universal base, as it can interact with each of the standard bases via two hydrogen bonds (Figure 1). The interactions in order of decreasing stability are I-C > I-A > I-T ≈ I-G.1 The first three pairs are standard anti-anti interactions, whereas the last pair is an unusual I(syn) – G(anti) interaction.
 |
 |
 |
 |
|
Figure 1. Base pairing interactions of inosine. |
Due to these base pairing properties, inosine has been used in a number of applications predominantly involving either synthetic primers or probes.2, 3 In cases where the target position is ambiguous, an inosine can be used to maximize hybridization as an alternative to degenerate oligonucleotides. This is particularly applicable when many ambiguous positions are present. A somewhat recent example of this is the use of inosine in 16S rRNA sequencing.4
16S rRNA sequencing is a standard method for the taxonomic identification of bacteria. The technique involves the use of PCR to generate an amplicon of a highly conserved region. The amplicon is then sequenced, and the resulting data is compared with public databases. Ben-Dov et al. used this technique to analyze the different types of bacteria present in an industrial waste water evaporation pond. The analysis was carried out with the common primer pair of 8F and 907R, as well as a variation of the same primer pair in which the 3’-terminal residues were replaced with 2’-deoxyinosine, 8F-I, and 907R-I, respectively. Sequencing data revealed that the diversity obtained from the 8F-I/907R-I pair was close to twice that of the 8F/907R pair. Interestingly, although the native primer pair gave less diversity, it did have results for one phylum of bacteria that was absent from the 8F-I/907R-I results, suggesting that the use of both primers with and without inosine may be the superior method in analyzing very diverse microbial samples.
Figure 2. 2’-Fluoro-Inosine-CE Phosphoramidite
To complement our existing DNA, RNA, and 2’-OMe versions of inosine, we are adding the 2’-F version (Figure 2). This phosphoramidite requires a coupling time of 3 min, and synthesized inosine-containing oligonucleotides can be deprotected with standard conditions. As is generally the case for 2’-F RNA, heating in AMA will lead to some degradation and should be avoided.
References