In our December 2011 Glen Report 23.2, we introduced AminoOxy-Modifier-11 CE Phosphoramidite (1) as a convenient reagent for labelling DNA with aldehydes to form stable oxime-labelled oligonucleotides. Historically, oxime conjugations are performed under mildly acidic conditions, pH 4.5, to catalyze the reaction for rapid conjugation.1 At pH 7.4, however, the oxime conjugation can be much slower and a catalyst may be required to achieve a reasonable rate of reaction. Aniline has been used as a catalyst for oxime conjugations and is typically used at significant excess to achieve reasonable effects.2
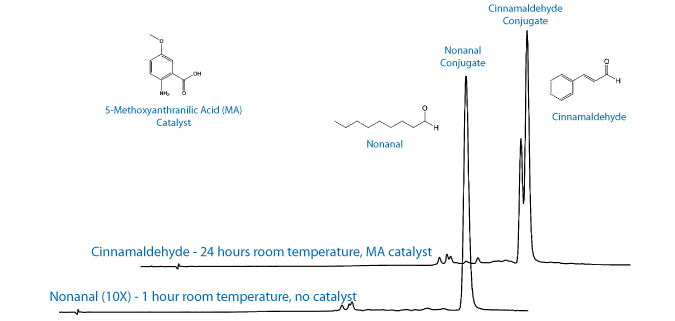
Recent work by the Kool group at Stanford has identified alternative catalysts that can be used at lower concentrations and still achieve a reasonable reaction rate.3,4 Of the catalysts reviewed by the Kool group, 5-methoxyanthranilic acid (5MA) was the most promising as it is soluble in aqueous buffer at pH 7.4, it is commercially available, and it is relatively inexpensive. Interestingly, they also report that the choice of aldehyde can have a significant effect on the overall rate of reaction. We chose to evaluate two different aldehydes, cinnamaldehyde and nonanal, for conjugation efficiency in the presence or absence of the catalyst. The results and experimental conditions are shown in Table 1.
| Room Temperature |
Nonanal No Catalyst |
Nonanal With Catalyst |
Nonanal (10X) No Catalyst |
Cinnamaldehyde No Catalyst |
Cinnamaldehyde Catalyst |
|---|---|---|---|---|---|
| 1 hour | 82.0% | 85.7% | 94.6% | 3.4% | 12.5% |
| 2 hours | 84.9% | 86.2% | 94.9% | 10.4% | 26.1% |
| 5.5 hours | 88.5% | 88.0% | 94.2% | 22.1% | 54.6% |
| 24 hours | 92.7% | 92.2% | 94.5% | 72.4% | 94.6% |
Prepare a 125 mM stock solution of 5MA in PBS, adjusted to pH 7.4 using 2M aqueous sodium hydroxide. (It should be noted that the 5MA is poorly soluble at acidic pH but is fully soluble at pH 7.4.) Add 0.125 mmoles of 5'-aminooxy-modified DNA in PBS and 0.625 mmoles of aldehyde in DMSO/Water (1:1) to a reaction tube. For the reactions with no catalyst, dilute to 0.4 mL with PBS. For the reactions with catalyst, add 3.125 mmoles of catalyst and dilute to 0.4 mL with PBS. Sample 60 µL and dilute to 150 µL with water. Load onto a prepared Glen Gel-Pak™ 0.2 cartridge and pre-elute with 200µL of water. Elute with 300µL of water and inject directly on HPLC.
The choice of aldehyde, alkyl versus aryl, can have a significant effect on the overall rate of reaction. This is consistent with previous reports that the equilibration concentration of the aldehyde/ aminooxy addition complex along with the rate of dehydration determine the overall rate of reaction.1 In one hour without catalyst, the nonanal labelling reaction was 82% complete compared to 3% labelling with cinnamaldehyde. In one hour with catalyst, the nonanal oligo was 86% labelled in 1 hour compared to 13% labelled for cinnamaldehyde. At higher concentrations of nonanal (10 equivalents) the labelling reaction is essentially complete (95%) in one hour, whereas cinnamaldehyde required 24 hours for complete reaction.
These results indicate that fluorophores and other aldehyde labels for oxime conjugations should have an alkyl linker to dramatically improve the kinetics of the reaction. This may have practical benefits for intracellular labelling, in situ labelling, and surface labelling where high concentrations of catalyst may not be feasible yet high labelling efficiency is required. For general oxime conjugations, the catalysts aniline and 5-methoxyanthranilic acid can be used to reduce the time and increase the overall efficiency of the labelling reactions.
For aryl aldehydes, we recommend 5-10 equivalents of aldehyde, 25 equivalents of 5MA, and reaction overnight at room temperature. For alkyl aldehydes, we recommend 10 equivalents of aldehyde and a 2 hour reaction at room temperature.
In summary, oxime labelling offers a mild, efficient, aqueous method for the labelling of oligonucleotides at acidic and neutral pH. The oxime is stable to standard oligonucleotide deprotection conditions and the reaction is bioorthogonal. Conjugation efficiency is based on the structure of the aldehyde, pH, and the presence of catalyst.