
Vijay Singh and Eric T. Kool
Department of Chemistry
Stanford University
Stanford, CA 94305 (USA)
The strategy of small-molecule fluorescent labeling of genetically encoded proteins has become a popular alternative to GFP labeling. This is because the methodology offers the advantage of much greater temporal resolution as the dye can be added at any point during the cell cycle or stage of an organism’s development.
Among the most widely used approaches is the HaloTag method developed by Promega, which utilizes a bacterial haloalkane dehalogenase. The enzyme removes halides from aliphatic hydrocarbons by a nucleophilic displacement mechanism to form a covalent ester linkage between the haloalkane and Asp106 in the enzyme. In the wild type haloalkane dehalogenase, the ester is quickly hydrolyzed by histidine 272 in the catalytic active site. However, by mutating the histidine to phenylalanine, the HaloTag variant renders the covalent ester bond stable toward hydrolysis, as shown in Figure 1.
The HaloTag domain is an exceedingly simple way to form covalent bonds between a variety of different molecules and labels to essentially any protein of interest.1 Standard ligands for HaloTag protein use lengthy chloroalkyl-substituted groups. Previously, we described a method for conjugating oligonucleotides to proteins by employing a specific phosphoramidite carrying a chloroalkyl linker which required several steps to prepare.2
To avoid these synthetic steps, we considered the possibility that a 5’-Bromohexyl Phosphoramidite could be used in combination with the Spacer Phosphoramidite 18 (a hexaethyleneglycol linker), both of which are commercially available from Glen Research, to produce a substrate for the HaloTag domain. We carried out preliminary experiments using the sequence, 5’-Bromohexyl-Spacer 18-(dT)15-Fluorescein-3’ to provide a fluorescent label for visualization.
 |
 |
Oligonucleotides with and without bromohexyl linkers were tested for HaloTag protein labeling using a GST-HaloTag fusion protein in vitro, with the control oligo using a 5’-Hexynyl linker. The GST-HaloTag fusion protein (0.5 μg (1.0 μM)) was treated with the oligonucleotides at 5.0 μM in PBS at 37°C for 1h. The reaction mixture was then resolved by SDS-PAGE and analyzed by fluorescence gel imaging (Ex 488 nm). The labeling experiment showed robust HaloTag protein labeling only with the oligonucleotide having the bromohexyl linker; we observed no protein labeling with the control oligo having no bromohexyl linker (see Figure 2 A).
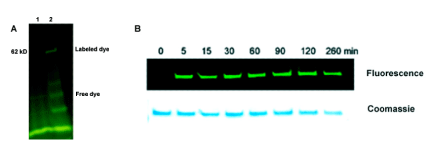
Next we evaluated the efficiency of protein labeling with the bromohexyl linker. In a time course experiment, 0.5 μg (1.0 μM) GST HaloTag fusion protein was incubated with 5.0 μM bromohexyl-modified oligo in PBS at 37°C and reaction was stopped at different time intervals and evaluated by gel with fluorescence imaging. The data showed rapid labeling of GST-HaloTag fusion protein with the 5’-Bromohexyl-modified oligo, with reaction apparently complete within 5 minutes (see Figure 2 B). Thus, the results from this initial test indicate the GST-HaloTag fusion protein is rapidly and efficiently conjugated to an oligo employing the 5’-Bromohexyl Phosphoramidite in combination with a hexaethylene glycol linker at the 5’ end of an oligonucleotide.
See Preparation of Bromohexyl-modified oligonucleotides for HaloTag conjugation