Glen Report 6.23: Research Review - Antisense RNA
The synthesis of 2'-OMe-RNA is as simple to perform as DNA synthesis and avoids most of the problems associated with RNA synthesis. Oligo-2'-OMe-nucleotides are resistant (but not totally resistant) to a variety of ribo- and deoxyribonucleases. As well as being stable to normal handling and nuclease resistant, oligo-2'-OMe-nucleotides form more stable hybrids with complementary RNA strands than equivalent DNA and RNA sequences. This combination of useful properties promises to make 2'-OMe-RNA sequences powerful research tools as has been noted in a recent review1.
Antisense Activities
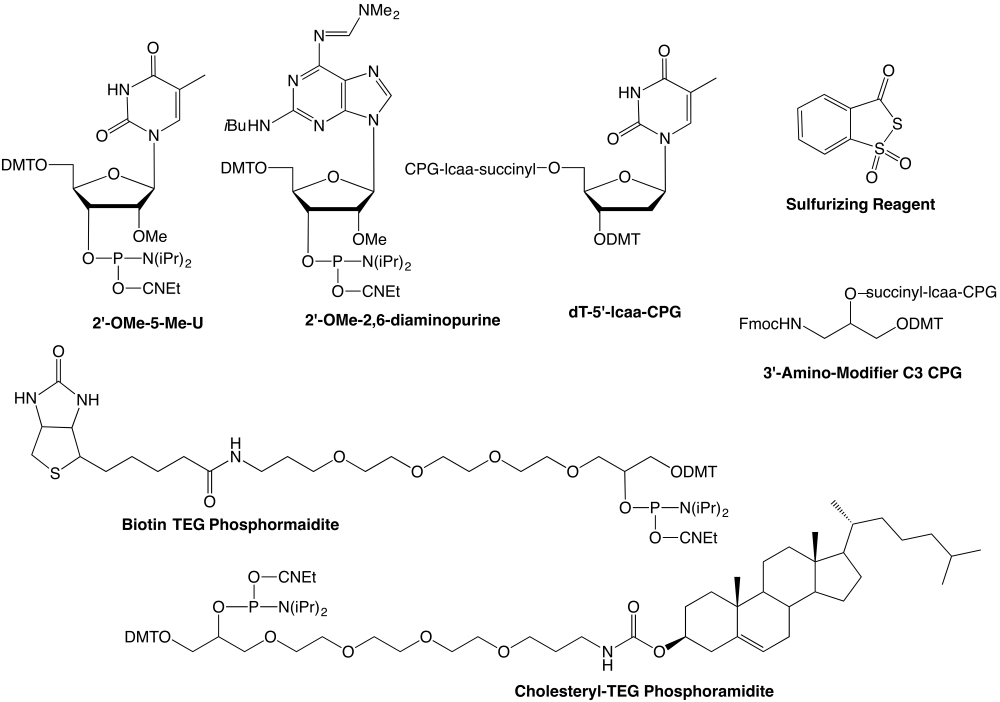
Oligonucleotides are believed to induce cleavage of RNA by RNase H, thereby reducing levels of the target RNA. Although 2'-OMe-RNA sequences exhibit a high affinity for RNA targets, these sequences are not substrates for RNase H. The combination of requirements of nuclease resistance, high affinity for target RNA, and being substrates for RNase H, has led to the evaluation of chimeric oligo-2'-OMe-nucleotides containing oligodeoxynucleotide gaps as antisense therapeutics. A group at Isis Pharmaceuticals has described2 chimeric antisense oligonucleotides based on 2'-modified oligonucleotides containing oligodeoxynucleotide gaps with all internucleotide linkages modified to phosphorothioates for even better nuclease resistance. The results indicate that in the system tested, a minimum of five oligodeoxynucleotide linkages in the center are necessary for maintaining RNase H and antisense activity. An additional route to enhancing exonuclease resistance of antisense oligonucleotides is to invert the nucleoside at the 3'-terminus3 with a 3'-3' linkage. This is readily accomplished using an appropriate support in which the nucleoside is attached using the 5'-hydroxyl and synthesis proceeds from the 3'-hydroxyl group. Eventual digestion of the modified oligonucleotide leads to only natural nucleosides. Similarly, the 3'-terminus can be blocked with an aminoalkyl group4.
Enhanced Binding
In situations where higher affinity for the target strand is sought, it is possible to substitute A sites with 2-amino-A (2,6-diaminopurine) which sets up an additional hydrogen bond to U. The Tm of the hybrid forms increases by 3¡ for every site modified1. This is especially important in cases where the target is U-rich5. Enhanced binding can be achieved by the simple substitution of Thymidine for U since the additional 5-methyl group leads to a more stable duplex. A further instance where substitution of a modified base to enhance duplex stabilization is found in the demonstration6 that 5-propynyl-U and 5-propynyl-C (as the 2'-O-allyl derivatives) exhibit a higher affinity for the target RNA strand. Each U substitution enhances Tm by 0.3¡ while each C substitution enhances Tm by 0.7¡. (This strategy is described in further detail beginning on the front page of this issue.)
Enhanced Cell Penetration
Transport of oligonucleotides into cells is a problem routinely faced by antisense researchers. This process would be expected to be improved by modifying the antisense oligonucleotides with suitable lipids. Recently, enhanced penetration of oligo-2'-OMe-nucleotides modified by the attachment of cholesterol (via a disulfide linkage) into liposomes was reported7. The attachment of cholesterol improved both affinity for and internalization of the oligo-2'-OMe-nucleotides.
Antisense Probes
The combination of properties offered by oligo-2'-OMe-nucleotides - nuclease resistance, stable hybrid formation, ease of synthesis - make them especially suitable for use as antisense probes. In this mode, RNase H resistant oligo-2'-OMe-nucleotides, labelled or unlabelled, can be used to block specific RNA functions8 and, when attached to a suitable tag, usually biotin9, can be used in affinity purification of RNA complexes. These activities are reviewed in detail by Lamond and Sproat1 whose contribution to research in this area has been especially noteworthy.
References:
(1) A.I. Lamond and B.S. Sproat, FEBS Letters, 1993, 325, 123-127.
(2) B.P. Monia, E.A. Lesnik, C. Gonzalez, W.F. Lima, D. Mcgee, C.J. Guinosso, A.M. Kawasaki, P.D. Cook, and S.M. Freier, J. Biol. Chem., 1993, 268, 14514-14522.
(3) J.F.R. Ortigao, H. Rosch, H. Selter, A. Frohlich, A. Lorenz, M. Montenarh, and H. Seliger, Antisense Res. & Dev., 1992, 2, 129-146.
(4) J.G. Zendegui, K.M. Vasquez, J.H. Tinsley, D.J. Kessler, and M.E. Hogan, Nucleic Acids Res., 1992, 20, 307-314.
(5) G.M. Lamm, B.J. Blencowe, B.S. Sproat, A.M. Iribarren, U. Ryder, and A.I. Lamond, Nucleic Acids Res., 1991, 19, 3193-3198.
(6) B.C. Froehler, R.J. Jones, X.D. Cao, and T.J. Terhorst, Tetrahedron Lett., 1993, 34, 1003-1006.
(7) B. Oberhauser and E. Wagner, Nucleic Acids Res., 1992, 20, 533-538.
(8) M. Cotten, B. Oberhauser, H. Brunar, A. Holzner, G. Issakides, C.R. Noe, G. Schaffner, E. Wagner, and M.L. Birnstiel, Nucleic Acids Res., 1991, 19, 2629-2635.
(9) H.O. Smith, K. Tabiti, G. Schaffner, D. Soldati, U. Albrecht, and M.L. Birnstiel, Proc. Nat. Acad. Sc. USA, 1991, 88, 9784-9788.
Product Information
2'-OMe RNA Synthesis
Sulfurizing Reagent (40-4036) has been discontinued. Please see:
Sulfurizing Reagent II (40-4037)
dT-5'-CPG (20-0302)
3'-Amino-Modifier C3 CPG (20-2950) has been discontinued. Please see:
3'-PT-Amino-Modifier C3 CPG (20-2954)
2'-OMe-2,6-Diaminopurine-CE Phosphoramidite (10-3124)
Cholesteryl-TEG Phosphoramidie (10-1975)
BiotinTEG Phosphoramidite (10-1955)

