Glen Report 23.21: Glen Research Epigenetic Bases Report
M. Mueller and T. Carell
Center for Integrated Protein Science (CiPSM) Department of Chemistry
Ludwig-Maximilians-University Butenandtstr. 5–13
81377 Munich, Germany
Heritable control of gene expression is the field of epigenetic research. DNA and histone modifications can switch a gene from its ‘on’ to the ‘off’ state and vice versa. Until 2009, the knowledge about DNA modifications was limited to the introduction of a methyl group at C5 of the cytosine base, which converts 2’-deoxyCytidine (dC) into 5-methyl-2’-deoxyCytidine (mdC). (See Figure 1 for structures of the modified pyrimidine bases discussed in this article.) This methylation occurs predominantly in CpG islands (areas with high occurrence of the CG motif) of promoters. mdC is introduced by specialized DNA methyltransferase (DNMT) enzymes.
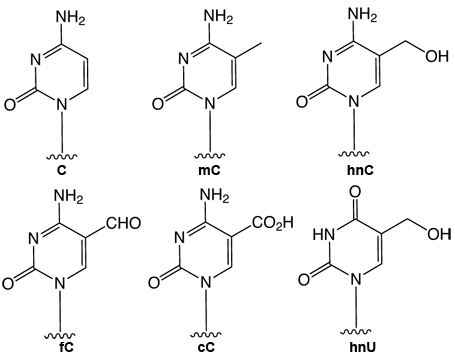
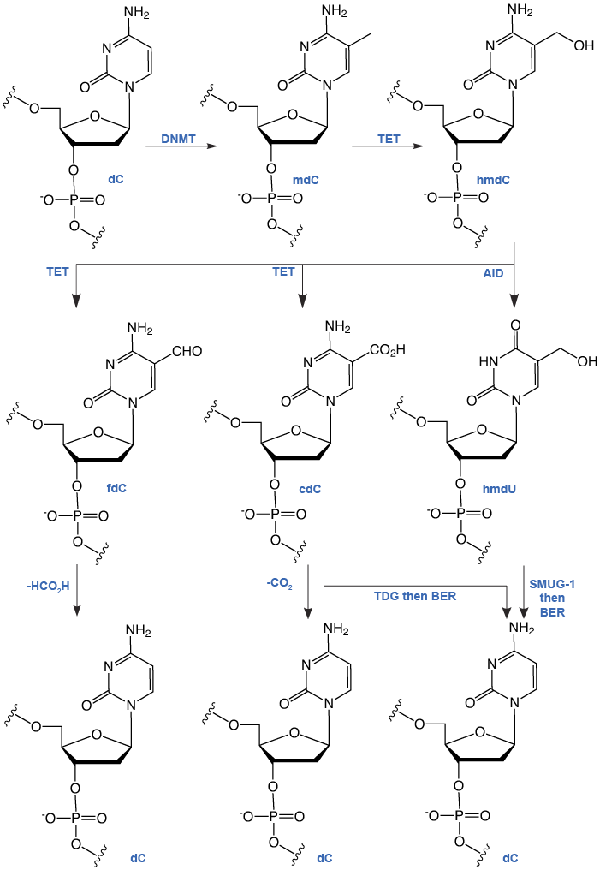
In 2009, two reports1,2 described the discovery of 5-hydroxymethyl-2’-deoxyCytidine (hmdC), a novel dC modification in Purkinje neurons and embryonic stem cells. Later, a third report found this modification to be strongly enriched in brain tissues associated with higher cognitive functions.3 This new dC modification is generated by the action of a-ketoglutarate dependent TET enzymes (ten eleven translocation), which oxidizes mdC to hmdC, as shown in Figure 2. This finding stimulated discussion about active demethylation pathways that could occur, e.g., via base excision repair (BER), with the help of specialized DNA glycosylases. Alternatively, one could envision a process in which the hydroxymethyl group of hmdC is further oxidized to a formyl or carboxyl functionality followed by elimination of either formic acid or carbon dioxide4,5 (Figure 2).
A number of recent publications provides data that support both pathways. It was discovered that hmdC could be deaminated by activation-induced deaminase (AID) enzymes to provide 5-hydroxymethyl-2’-deoxyUridine (hmdU). This compound was shown to be excised by the SMUG-1 DNA glycosylase6 (Figure 2). After initial failure to detect any further oxidized hmdC derivatives in somatic tissues4, newly developed mass spectrometric technologies, in combination with the available reference compounds, finally enabled researchers to gather strong support for the putative oxidative demethylation pathway. These methods and standards enabled the discovery of 5-formyl-2’-deoxyCytidine (fdC) in differentiating embryonic stem cells.7 Recently, a similar technology also led to the discovery of 5-carboxyl-2’-deoxyCytidine (cdC)8,9, but the amount of fdC and cdC measured differs largely in all three reports.
Research is currently ongoing to unravel the true levels and fate of these further oxidized dC bases in somatic tissues and in different stem cells. Even along the oxidative pathway, base excision processes have been proposed to play a major role with two reports showing that thymidine DNA glycosylase (TDG) accepts both fdC and cdC as substrates.8,10 A possible oxidative demethylation pathway would clearly rely on the existence of a dedicated decarboxylase that is able to convert cdC back into dC. Such an intriguing decarboxylation would enable nature to remove the 5-methyl group in mdC without introducing DNA strand-breaks that accompany any BER based base removal (Figure 2).
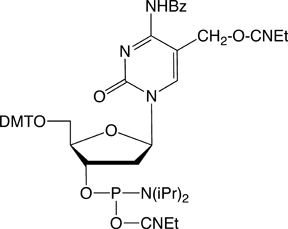
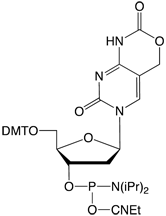


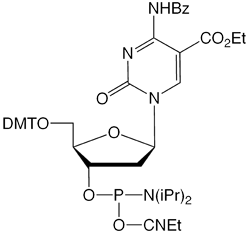
Deprotection conditions for all the phosphoramidite monomers included in this report are included in Table 1.
| Building Block*/Deprotection Conditions | NH4OH or AMA | NaOH | K2CO3 |
|---|---|---|---|
| 5-Hydroxymethyl-dC (1) | Ammonium hydroxide at 75°C for 17 hours. | Not compatible | Not compatible |
| 5-Hydroxymethyl-dC II (2) | Yields urea derivatives – possibility to introduce different ureas post-synthetically by deprotection with suitable amine | 0.4 M NaOH in methanol/water 4:1, 25°C, 17h (NaOH deprotection is not compatible with DMF protecting groups) |
0.05M K2CO3 in anhydrous methanol, 25°C, 17h using UltraMild conditions |
| 5-Formyl-dC (3) | Standard deprotection followed by periodate oxidation, see Figure 4 | Standard deprotection followed by periodate oxidation, see Figure 4 | Standard deprotection followed by periodate oxidation, see Figure 4 |
| 5-Formyl-dC II (4) | Conc. NH5OH, 25°C, 17h Initially yields imine, which is hydrolysed by water |
0.4 M NaOH in methanol/water 4:1, 25°C, 17h (NaOH deprotection is not compatible with DMF protecting groups) |
Benzoyl protecting group is not compatible with UltraMild deprotection conditions |
| 5-Carboxy-dC (5) | Yields amides as well as desired carboxylic acid | 0.4 M NaOH in methanol/water 4:1, 25°C, 17h (NaOH deprotection is not compatible with DMF protecting groups) |
Not compatible |
| *See Figure 3 for structures | |||
References
- S. Kriaucionis, and N. Heintz, Science, 2009, 324, 929-30.
- M. Tahiliani, et al., Science, 2009, 324, 930-935.
- M. Münzel, et al., Angewandte Chemie-International Edition, 2010, 49, 5375-5377.
- D. Globisch, et al., PLoS One, 2010, 5, e15367.
- S.C. Wu, and Y. Zhang, Nat Rev Mol Cell Biol, 2010, 11, 607-20.
- J.U. Guo, Y.J. Su, C. Zhong, G.L. Ming, and H.J. Song, Cell, 2011, 145, 423-434.
- T. Pfaffeneder, et al., Angewandte Chemie-International Edition, 2011, 50, 7008-7012.
- Y.F. He, et al., Science, 2011, 333, 1303-1307.
- S. Ito, et al., Science, 2011, 333, 1300-1303.
- A. Maiti, and A.C. Drohat, J Biol Chem, 2011, 286, 35334-8.
- M. Münzel, D. Globisch, C. Trindler, and T. Carell, Org Lett, 2010, 12, 5671-3.
- M. Münzel, et al., Improved Synthesis and Evaluation of Oligonucleotides Containing 5-Hydroxymethylcytosine, 5-Formylcytosine and 5-Carboxylcytosine. In Chemistry - A European Journal, 2011, in press.
- N. Karino, Y. Ueno, and A. Matsuda, Nucleic Acids Res., 2001, 29, 2456-63.
Product Information
- Glen Report 23.21: Glen Research Epigenetic Bases Report
- Glen Report 23.22: New Product – 5’-AminoOxy-Modifier 11
- Glen Report 23.23: New Products - BlackBerry® Quencher (BBQ-650®)
- Glen Report 23.24: On-resin Synthesis of Maleimido-oligonucleotides
- Glen Report 23.25: New Product - NPOM-Caged-dT
- Glen Report 23.26: Glen-Pak™ Purification - Then and Now
- Glen Report 23.27: Technical Brief - Deprotection of HMdU
- Glen Report 23.28:Technical Brief - Improved Oligo Synthesis using 2-Amino-dA and CSO Oxidation
- Glen Report 23.29: New Products - Bulk Polystyrene Supports

