Glen Report 16.26: Still More New Products
PC Linker Phosphoramidite
Photo-triggered DNA cleavage is a major tool used for studying conformational changes and strand breaks, as well as for studying activation of nucleic-acid-targeted drugs, such as antisense oligonucleotides. A versatile photocleavable DNA building block has been described by researchers in Washington University, Missouri and used in phototriggered hybridization.1 In association with Link Technologies Ltd (Scotland, ) this phosphoramidite is now available from Glen Research.
This reagent has also been used in the design of multifunctional DNA and RNA conjugates2 for the in vitro selection of new molecules catalyzing biomolecular reactions. Researchers at Bruker Daltonik in Germany have also developed genoSNIP, a method for single-nucleotide polymorphism (SNP) genotyping by MALDI-TOF mass spectrometry.3 This method uses size reduction of primer extension products by incorporation of the photocleavable linker for phototriggering strand breaks near to the 3' end of the extension primer.
Similar to the 3 other available PC monomers, the general design of the PC Linker is based on an a-substituted 2-nitrobenzyl group. The photo-reactive group is derivatized as a β-cyanoethyl phosphoramidite and the non-nucleoside PC Linker can be incorporated into oligonucleotides at any position by standard automated DNA synthesis methodology. Coupling efficiencies >95% are achieved using an extended coupling time of 15 minutes. For ease of use, the product contains a dimethoxytrityl group. The β-cyanoethyl group is removed under normal synthesis conditions using 25% aqueous ammonia solution.
Upon irradiating a PC-modified oligo with near-UV light, the phosphodiester bond between the linker and the phosphate is cleaved, resulting in the formation of a 5’-monophosphate on the released oligonucleotide. Unlike other photocleavable spacers, the PC Linker Phosphoramidite has the added advantage in that photocleavage results in monophosphate fragments at both the 3’- and 5’-termini (see Figure 2).
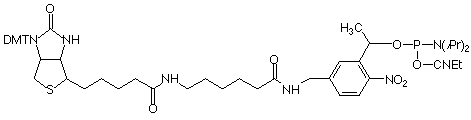
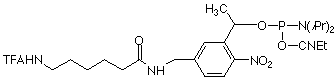
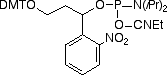
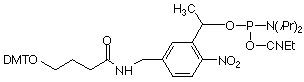

References
- P. Ordoukhanian and J-S. Taylor, J. Am. Chem. Soc., 117, 9570-9571, 1995.
-
- F. Hausch and A. Jäschke, Nucleic Acids Research, 2000, 28, e35.
- F. Hausch and A. Jäschke, Tetrahedron, 2001, 57, 1261-1268.
- T. Wenzel, T. Elssner, K. Fahr, J. Bimmler, S. Richter, I. Thomas, and M. Kostrzewa, Nucleosides, Nucleotides & Nucleic Acids, 2003, 22, 1579-1581.
Product Information
Photocleavable Phosphoramidites
DesthiobiotinTEG
Avidin, streptavidin and other biotin-binding proteins have the ability to form an intense association with biotin-containing molecules. This association has been used for many years to develop systems designed to capture biotinylated biomolecules. In the oligonucleotide field, probably the most common modification is biotinylation and reagents are available to modify oligos at the termini and within the sequence. A wide variety of tests and techniques are in routine use to exploit the extraordinary affinity of these biotin-binding proteins for biotinylated biomolecules. However, the intense affinity of biotin-binding proteins for biotin is also the biggest drawback in that the association is essentially irreversible. Indeed, extremely low pH and highly concentrated chaotropic reagents are required to break the association and these conditions are not entirely compatible with oligonucleotides. 2-Iminobiotin has been used as a reversibly binding biotin reagent since its association with biotin-binding proteins can be broken at ph4. However, 2-iminobiotin is not stable to the conditions of oligonucleotide deprotection. Another biotin analogue that exhibits lower binding to biotin-binding proteins like streptavidin is desthiobiotin (or dethiobiotin). This biotin analogue is lacking the sulfur group from the molecule and has a dissociation constant (Kd) several orders of magnitude less than biotin/streptavidin. As a result, biomolecules containing desthiobiotin are dissociated from streptavidin simply in the presence of buffered solutions of biotin.1 We believe that these characteristics will allow another biotin product to prosper.
Our most versatile biotin products are the biotinTEG products which can be added singly or in multiple additions anywhere within an oligonucleotide. In addition, the triethyleneglycol (TEG) section of the structure separates the biotin from the oligonucleotide in such a way that it is more readily captured by streptavidin. We have used the same structure to offer desthiobiotinTEG phosphoramidite and the corresponding CPG.
Reference
- J.D. Hirsch, L. Eslamizar, B.J. Filanoski, N. Malekzadeh, R.P. Haugland, and J.M. Beechem, Anal Biochem, 2002, 308, 343-57.
Product Information
DesthiobiotinTEG Phosphoramidite (10-1952)
DesthiobiotinTEG-CPG (20-2952)
5-Me-2'-deoxyZebularine
Zebularine (pyrimidin-2-one ribo-nucleoside) (1) is a cytidine analog that acts as a DNA demethylase inhibitor, as well as a cytidine deaminase inhibitor. This structure is very active biologically and Zebularine is now used as a potent anti-cancer drug. A 2'-deoxynucleoside analogue of Zebularine, 5-methyl-pyrimidin-2-one, 2'-deoxy-nucleoside (2), has been used1 to probe the initiation of the cellular DNA repair process by making use of its mildly fluorescent properties. We believe that this combination of biological activity and fluorescence properties would make the phosphoramidite (3) a strong addition to our array of nucleoside analogue phosphoramidites.
Reference
- S.F. Singleton, et al., Org Lett, 2001, 3, 3919-22.
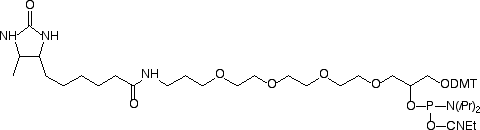
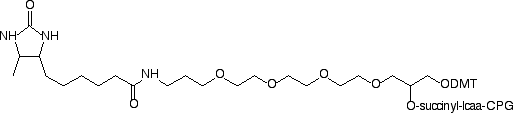



Product Information
- Glen Report 16.21: Thymine dimers - DNA Lesions Induced By Sunlight CIS-SYN Thymine Dimer Phosphoramidite Now Available
- Glen Report 16.22: EraGen BioSciences Technology (courtesy of EraGen Biosciences)
- Glen Report 16.23: Activators, Columns and Plates
- Glen Report 16.24: Locked Nucleic Acid (LNA™) Phosphoramidites
- Glen Report 16.25: Trimer (Codon) Phosphoramidites Simplify Library Preparation
- Glen Report 16.26: Still More New Products
- Glen Report 16.27: New Epoch Products - 5'-Aldehyde-Modifier C2 Phosphoramidite and Gig Harbor Green™ Phosphoramidite

