Glen Report 11.11: Using Modified Bases to Optimize Hybridization
Introduction
The vast majority of synthetic oligonucleotides today are destined for use as primers in PCR or in sequencing applications, and current systems - from primers to amplification techniques - seem to work very well indeed. Of course, optimization of base pairing is always desirable and, as long as the cost does not outweigh the benefits, may be the answer to some challenging research problems.
2-Amino-dA
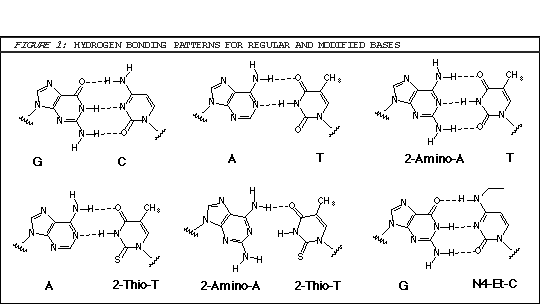
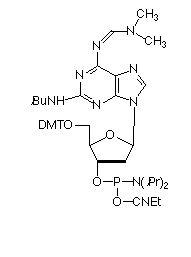
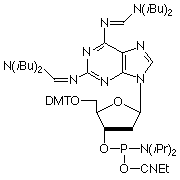
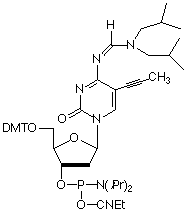
As shown in Figure 1, A-T base pairs have two hydrogen bonds whereas G-C base pairs have three hydrogen bonds. The simplest approach to improving primers would be to substitute A sites with 2-amino-A which forms three hydrogen bonds with T on hybridization.1 2-Amino-A also destabilizes A-G wobble mismatches, thus increasing specificity. Although 2-amino-dA monomers have been commercially available, they have had two severe drawbacks: the protection scheme and the cost. Because 2-amino-dA is very susceptible to depurination during the acidic deblocking step of DNA synthesis, mild deprotecting groups like PAC to protect both amino groups should not be used. The combination of N2-isobutyryl and N6-formamidine protecting groups in our earlier monomer (1) stabilized the monomer to depurination but made it very slow to deprotect, requiring 7 days in ammonium hydroxide at 55° or 17 hours at 55° in AMA for complete removal. After a significant development effort, we are happy to announce a new 2-amino-dA monomer (2) which appears to solve all of the earlier problems: deprotection is fast and effective in ammonium hydroxide; it is stabilized to depurination during synthesis; and the cost is only about 20% of the earlier monomer.
Pyrimidine Analogues
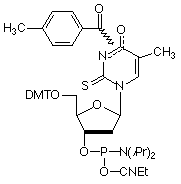
Substitution of 5-Me-dC1, 2 (3) or, better, C-5 propynyl-dC (4) for dC3 and C-5 propynyl-dU (5) for dT3 are effective strategies to enhance base pairing. This increase in hybridization efficiency is due to the hydrophobic nature of the groups at the C-5 position which helps to exclude water molecules from the duplex.
Duplex Stabilization
Using these base substitutions, duplex stability and therefore melting temperatures are raised by the approximate amounts shown below:
| Base substitution | Tm increase |
|---|---|
| 2-Amino-A | 3.0° per substitution |
| 5-Methyl-C | 1.3° per substitution |
| C-5 propynyl-C | 2.8° per substitution |
| C-5 propynyl-U | 1.7° per substitution |
While these modifications would also have a desirable effect on antisense oligonucleotides, the increased costs associated with most of them may limit their use. However, primers are less cost-sensitive because of the smaller scale, so the effects of the modified bases may be more generally useable. Potential improvements would include: the ability to use shorter oligos when sequence information is incomplete; higher melting temperatures, which should minimize the frequency of mutations; and enhanced binding, which should break any secondary structure in the target.
SBC Oligos
Selectively Binding Complementary (SBC) oligonucleotides4 have the unique property of being able to simultaneously bind to both the sense and antisense strands of a DNA or RNA duplex. This should make them extremely useful for investigating secondary structures such as Holliday junctions and other branching moieties. They may also prove useful as antisense agents where the mRNA target exhibits significant secondary structure. SBC oligos possess this unique ability because, although they exhibit high affinity for natural oligonucleotides, they show little affinity for other SBC oligos even of a complementary sequence.
Oligos in which A has been replaced with 2-amino-A and T with 2-thio-T represent an excellent example of SBC oligos.4 While 2-amino-A forms a very stable base pair with T containing three hydrogen bonds, the stability of the base pair with 2-thio-T is greatly diminished. Model building suggests that steric interactions between the 2-thio group of thymidine and the 2-amino group of adenine tilt the bases relative to each other yielding a base pair that contains only a single hydrogen bond.4 However, 2-thio-T base pairs perfectly well with A, as shown in Figure 1. But the real proof is not in models: SBC 20mers annealed against a DNA 20mer target exhibited Tm values 10° higher than the corresponding DNA-DNA hybrid, whereas the SBC-SBC hybrid yielded Tm values 30° lower.4
Because of the resistance to hydrolysis of the protecting groups of our earlier 2-amino-dA monomer, the aggressive deprotection was not compatible with the presence of 2-thio-dT in the same oligo. Fortunately, our new 2-amino-dA monomer is rapidly deprotected in ammonium hydroxide and is compatible with 2-thio-dT (6). Now SBC oligos containing 2-amino-dA and 2-thio-dT can be easily prepared.
Hybridization Independent of Base Composition
Any technique that involves hybridization of multiple sequences simultaneously, as in DNA chip and reverse hybridization technologies, is subject to inaccuracies due to differences in GC content. Sequences with high GC content may contain mismatches and still hybridize, whereas a low GC content probe may match perfectly and yet disassociate from the target, leading to false positives and negatives respectively.
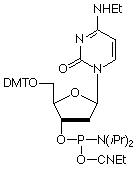
An elegant way of circumventing this problem would be to use a modified base that normalized the stability of the GC and AT base pairs. With this goal in mind, a series of modified dC bases was evaluated to develop a system where hybridization was independent of base composition and only dependent upon oligonucleotide length.5 It was found that the N4-ethyl analogue (N4-Et-dC) hybridizes specifically to natural dG but the stability of the base pair is reduced to about the level of an AT base pair. In a series of probes whose GC content ranged from 0 to 100%, the range in Tm values when N4-Et-dC was used was only 7°; when dC was used, that range was 39°. We are happy to offer N4-Et-dC (7) for oligo synthesis.

References
- Y. Lebedev, et al., Genetic Analysis - Biomolecular Engineering, 1996, 13, 15-21.
- L.E. Xodo, G. Manzini, F. Quadrifoglio, G.A.v.d. Marel, and J.H.v. Boom, Nucleic Acids Res., 1991, 19, 5625-5631.
- B.C. Froehler, S. Wadwani, T.J. Terhorst, and S.R. Gerrard, Tetrahedron Lett., 1992, 33, 5307-5310.
- I.V. Kutyavin, R.L. Rhinehart, E.A. Lukhtanov, V.V. Gorn, R.B. Meyer, and H.B. Gamper, Biochemistry, 1996, 35, 11170-11176.
- H.K. Nguyen, P. Auffray, U. Asseline, D. Dupret, and N.T. Thuong, Nucleic Acids Res., 1997, 25, 3059-65.
Product Information
2-Amino-dA-CE Phosphoramidite (10-1085)
5-Me-dC-CE Phosphoramidite (10-1060)
pdC-CE Phosphoramidite (10-1014)
pdU-CE Phosphoramidite (10-1054)
2-Thio-dT-CE Phosphoramidite (10-1036)
N4-Et-dC-CE Phosphoramidite (10-1068)
- Glen Report 11.11: Using Modified Bases to Optimize Hybridization
- Glen Report 11.12: Oligo Affinity Supports
- Glen Report 11.13: New Fluorescent Reagents - TAMRA-dT, Dabcyl-dT
- Glen Report 11.14: The Use of Puromycin CPG in Combinatorial Chemistry
- Glen Report 11.15: Update - Universal Supports, Q-supports
- Glen Report 11.16: More Novel Monomers - 6-Thio-dG, 2'-Amino-RNA, 5-Hydroxy-dC,dU Spacer C18, Spacer 9-CPG

