Glen Report 27.21: New Product - 1-Ethynyl-dSpacer CE Phosphoramidite
Introduction
Efficient post synthesis modification of DNA and RNA offers the capability to attach a diverse set of labels that would not otherwise be accessible by standard phosphoramidite oligonucleotide synthesis. Through copper (I) alkyne-azide cycloaddition (CuAAC), a variety of azide labels can be efficiently attached to alkyne-modified DNA and RNA oligonucleotides.1,2 As shown in Figure 1, our alkyne modifiers include internal nucleobase modifiers, 5'-modifiers, 3'-modifier supports, and serinol linkers.
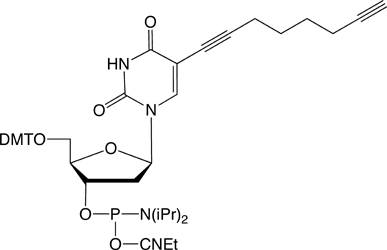
C8-Alkyne-dT |
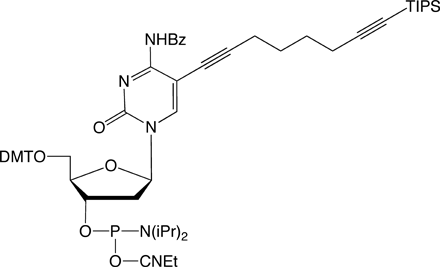
C8-TIPS-Alkyne-dC |
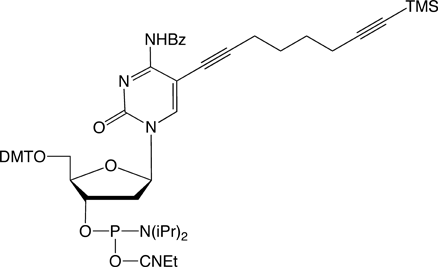
C8-TMS-Alkyne-dC |
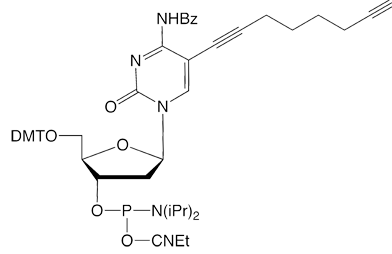
C8-Alkyne-dC |
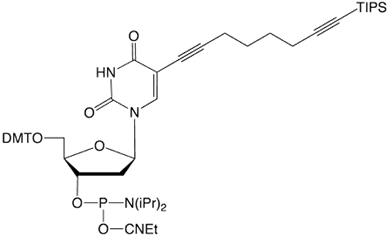
C8-TIPS-Alkyne-dT |
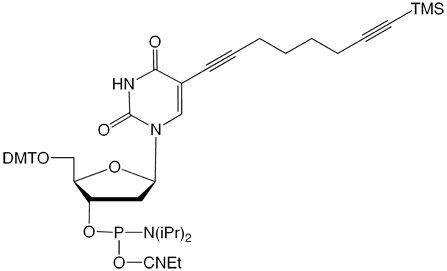
C8-TMS-Alkyne-dT |
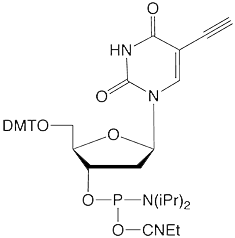
5-Ethynyl-dU |
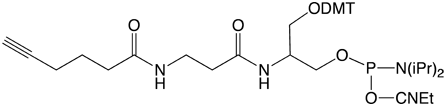
Alkyne-Modifier Serinol Phosphoramidite |
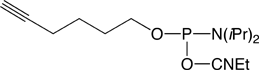
5’-Hexynyl Phosphoramidite |
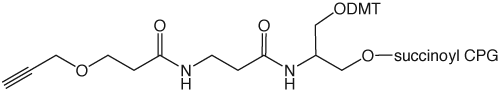
3’-Alkyne-Modifier Serinol CPG |
Figure 1: Structures of Glen Research Alkyne Modifiers |
|
1-Ethynyl-dSpacer
In this report, we introduce 1-Ethynyl-dSpacer CE Phosphoramidite, (1) in Figure 2, that can be used in any position within an oligonucleotide while still retaining the high efficiency of click chemistry.
The click reaction of this modifier with an azide forms a 1,2,3-triazol-4-yl pseudo-nucleobase structure, as shown in Figure 2. These pseudo-nucleobases can act as fleximers that have the potential to moderate duplex stability and enzymatic activities.3,4 In recent work, the 1,2,3-triazolyl modified DNA was evaluated for duplex stability in dsDNA and in triplex forming oligonucleotides (TFOs).5,6 Several modifications retained duplex or triplex stability compared to the unmodified oligonucleotides and had the potential to function as universal bases.

Reaction of 1-Ethynyl-dSpacer with Psoralen Azide
Synthesis and Deprotection

Figure 2: 1-Ethynyl-dSpacer and its Click Reaction with Azides
The coupling efficiency of 1-ethynyl-dSpacer was optimal under the standard conditions used for regular DNA monomers.
Deprotection was evaluated for compatibility with the following conditions using a simple T6 oligo with a single addition of 1-ethynyl-dSpacer at the 5' terminus.
- Ammonium hydroxide, 55°C overnight
- Ammonium hydroxide, 2 hours, RT
- AMA, 65°C 10 minutes
- Potassium carbonate in methanol, 4 hours, RT
- Ammonium Hydroxide, 5 minutes, RT (control)
No degradation was observed under these deprotection conditions.
Each deprotected sample was clicked with HEX azide to evaluate the possibility of any degradation by hydration occurring during deprotection. None was observed as all oligos labelled efficiently.
To evaluate for compatibility with standard synthesis reagents, oligos were exposed to reagents for an equivalent of a 50-mer exposure. The oligos showed broad compatibility with standard reagents. All oligos were clicked with HEX azide after exposure to confirm there was no degradation caused by exposure to the reagents.
The modifier is efficiently incorporated into oligonucleotides by standard phosphoramidite chemistry, is stable to common deprotection conditions, and is compatible with Glen-Pak™ purification and produces very pure oligonucleotides. The best deprotection results were obtained with AMA for 10 minutes at 65°C.
Click Evaluation
The resulting modified oligonucleotide clicked efficiently using standard click chemistry with THPTA and a selection of azides (Figure 3). The high efficiency of the click reaction with a variety of azides is illustrated in Figure 4.

Figure 3: Azides used in this Study

Figure 4: Sample 5RP HPLC of Modified Oligos
Interestingly, the efficiency of the cycloaddition does not appear to be significantly affected by the azide tag. In a simple experiment, a library of azides was evaluated for its efficiency to click to a 1-ethynyl-dSpacer labelled oligonucleotide in a single reaction. Generally, a minimum of 4 equivalents of azide maximizes labelling while minimizing the reaction time. In this experiment, the total azide concentration was slightly over 1 equivalent. All expected modifications were observed in the library by HPLC and UV/Vis spectroscopy.
Duplex Stability
1-Ethynyl-dSpacer was incorporated into the T7 forward primer and clicked with several azides. The oligos were evaluated for their effect on duplex stability. The 1-ethynyl-dSpacer modification exhibits similar duplex stability to the standard dSpacer (10-1914) and destabilizes the duplex when internally incorporated. Upon cycloaddition, the duplex stability is moderated by the resulting structure of the modification. Simple 1,2,3-triazoles were destabilizing, as were modifications that incorporated TEG linkers (6-FAM-TEG and Amino-TEG). Modifications that incorporated aromatic functional groups restored duplex stability to varying degrees with coumarin restoring stability by 11°C and psoralen by 7°C. The results are shown in Table 1. Examples of the melting curves are shown in Figure 5.

Figure 5: Melting Curves using T7 Forward Primer and Complement
Conclusion
1-Ethynyl-dSpacer CE Phosphoramidite is a new tool in the click chemistry toolbox with the potential for simple access to a diverse set of modifications. As a dSpacer, the 1-ethynyl-dSpacer modifier has a similar profile to the current dSpacer, exhibiting the same stability to synthesis and deprotection with a similar effect in DNA duplexes. As an alkyne modifier, 1-ethynyl-dSpacer combines the flexibility of an internal or terminal modifier with the high efficiency of click chemistry. Additionally, 1-ethynyl-dSpacer is a unique modifier which generates a substituted 1,2,3-triazole pseudo-nucleobase after click chemistry. These modified pseudo-nucleobases have the potential for use in research as universal bases or hybridization probes.
| Modification | Tm |
|---|---|
| 1-Ethynyl-dSpacer | 40°C |
| 6-HEX | 40°C |
| 6-FAM-TEG | 40°C |
| Psoralen | 47°C |
| Coumarin | 51°C |
| Propanol | 43°C |
| Amino-TEG | 43°C |
| dSpacer (10-1914) | 40°C |
| Control, X=A | 55°C |
| T7 forward primer and complement 5’-TAA TAC GXC TCA CTA TAG GG - 3’ 5’-CCC TAT AGT GAG TCG TAT TA - 3’ |
|
Summary
- 1-Ethynyl-dSpacer is stable to standard synthesis conditions.
- No changes needed to standard coupling.
- The product is stable to standard deprotection conditions.
- The product can be used as an internal modification and a terminal modification.
- The product is stable to CuAAC and clicks efficiently.
- The product has similar duplex stability to dSpacer.
- 1-Ethynyl-dSpacer lowers the stability of the duplex by the same amount as dSpacer.
- Upon clicking, the duplex stability can be modified based on the resulting structure of the incorporation.
- The product has been evaluated with the following azides:
- 6-HEX Azide
- 6-FAM-TEG Azide
- Psoralen Azide
- Coumarin Azide
- Cyanine 7 Azide
- Azido propanol
- Amino-TEG Azide
References
- V.V. Rostovtsev, L.G. Green, V.V. Fokin, and K.B. Sharpless, Angew Chem Int Ed, 2002, 41, 2596-2599.
- Q. Wang, et al., Journal of the American Chemical Society, 2003, 125, 3192-3193.
- S.C. Zimmermann, et al., MedChemComm, 2011, 2, 650-654.
- A.H. St. Amant, L.A. Bean, J.P. Guthrie, and R.H.E. Hudson, Organic & Biomolecular Chemistry, 2012, 10, 6521-6525.
- M. Nakahara, et al., Bioorganic & Medicinal Chemistry Letters, 2009, 19, 3316-3319.
- Y. Hari, et al., Bioorganic & Medicinal Chemistry, 2011, 19, 1162-1166.
Ordering Information
- Glen Report 27.21: New Product - 1-Ethynyl-dSpacer CE Phosphoramidite
- Glen Report 27.22: Introducing 2’-OMe-Thiophosphoramidites
- Glen Report 27.23: New Product – Pac-2-Amino-dA
- Glen Report 27.24: New Product - 1-Methyl-Pseudouridine
- Glen Report 27.25: Technical Brief - Dual-labelled Oligos using Click Chemistry
- Glen Report 27.26: New Product - COT Serinol Phosphoramidite

