Glen Report 15.14: More and Still More Novel Minor Bases
What would a Glen Report be without a selection of new nucleoside analogues? Of course, GR 15.1 is no different!
7-deaza-8-aza-dG (PPG)
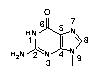
One of the perennial problems of DNA research occurs when analyzing DNA segments that are G-rich. Basically, DNA structure dictated by Watson-Crick base pairing is disrupted in G-rich segments because of their ability to create inter and intra strand hydrogen bonding. This aggregation causes enzymatic disruption so sequencing and PCR experiments become highly problematical. In probe experiments, these segments are not accessible due to this aggregation of G residues on the probe and/or target. The problem arises from extra hydrogen bonding forming at the N7 position of G (1).
Traditionally, 7-deaza-G (2) has been used to overcome these problems with some success. However, the 7-deaza-G - C base pair is destabilized relative to the G - C base pair by around 1° per insertion. Also, 7-deaza-G is relatively unstable to the iodine oxidation in the regular synthesis cycle and, if several insertions have to be made into an oligonucleotide, a non-iodine containing oxidizer must be used.
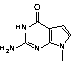
A solution to these problems may be found in the substitution of 7-deaza-8-aza-G (pyrazolo[3,4-d]pyrimidine) (PPG) (3) for G. This modification has been examined1 and 7-substituted analogues were evaluated several years ago by Seela's group. In 7-deaza-8-aza-G, the N7 and C8 atoms of G are flipped (Figure 1), allowing the modified base to retain the same electron density as the guanine ring system. The 7-deaza-8-aza-G - C base pair was found to be stabilized relative to G - C by around 1° per insertion. This stability enhancement has led to interest in the use of PPG in diagnostic probe applications.
The 7-deaza-8-aza-dG-CE Phosphoramidite monomer (4) is a very stable structure and, therefore, requires no changes from the regular synthesis cycles and deprotection procedures. It is made available as part of our distribution agreement with Epoch Biosciences, Inc., discussed in detail on the Front Page of this issue.
Product Information
7-Deaza-8-aza-dG-CE Phosphoramidite (PPG) (10-1073)
7-Deaza-8-aza-dA
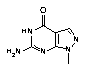
Similarly, the Adenine (5) analogue is 7-deaza-8-aza-A (6). Again, this molecule has been studied in depth over the years by Seela's group as the unsubstituted nucleoside2, but more recently with substituents on the 7-position. The melting behavior of 7-deaza-8-aza-A is similar to the G analogue in that the Tm of the 7-deaza-8-aza-A – T base pair is generally raised relative to the A - T base pair. The different electron density of the pyrazolo[3,4-d]pyrimidine ring system probably allows for better base stacking in a duplex.3
Again, the 7-deaza-8-aza-dA-CE Phosphoramidite monomer (7) is a stable structure and there is no need to change conditions during its use in oligonucleotide synthesis and deprotection.
Product Information
7-deaza-8-aza-dA-CE Phosphoramidite (10-1083)
2-F-dI
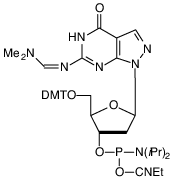
It has been a long time but we are at last adding to our list of Convertible Nucleosides, 2-F-dI-CE Phosphoramidite (8). 2-Fluoro-2'-deoxyInosine (2-F-dI) can be converted to 2-substituted dG derivatives by reaction with a primary amine, which displaces the fluorine atom.4,5 The timing of the conversion step is a little tricky because small alkyl primary amines are capable of doing the conversion while also cleaving and deprotecting the oligonucleotide. For example, reaction with ethylamine would convert 2-F-dI to N2-ethyl-dG but would simultaneously cleave and deprotect the oligonucleotide. Although that may be interesting in its own right, we have chosen to focus on larger primary amines in our development work. For example, treatment of the oligonucleotide (while still fully protected on the synthesis column) with dansyl cadaverine converts the nucleoside to an N2-dansyl-dG derivative, as shown in Figure 2. Further conventional deprotection of the oligonucleotide leads to the final product. The product oligonucleotide now has a fluorescent tag which, when hybridized to the target strand, will project into the minor groove of the double-stranded duplex. In a further example, we used cystamine to convert the 2-F-dI to a product containing a thiol group at the N2 position (Figure 2). Once this converted oligo is hybridized to the target, the thiol is available for cross-linking to, for example, a protein binding to the minor groove. The thiol can also form a disulfide crosslink with a similarly modified G on the complementary strand.6
As with all convertible nucleosides, we caution that these reactions are not trivial and should be undertaken by researchers with a good background in chemistry and access to appropriate analytical techniques.
.
Product Information
2-F-dI-CE Phosphoramidite (10-1082)
8-OMe-dG
Oxidative damage to G residues in biological systems leads to the formation of 8-oxo-G (9), the predominant product of G damage. 2-Aminoimidazolone (Iz, 10) and its hydrolysis product imidazolone (Z, 11) are also major oxidation products of G. Access to these two potential lesions is not possible during oligonucleotide synthesis because they are so base-labile. A suitable precursor, 8-methoxy-dG (8-OMe-dG, 12), to dIz has now been described.7 We have, therefore, added the convertible nucleoside monomer 8-OMe-dG CE Phosphoramidite (12) to our series of products offered for researching DNA damage and repair.
Oligonucleotide synthesis and deprotection in the presence of 8-OMe-dG is straightforward. The conversion of 8-OMe-dG to dIz takes place by irradiation of the oligonucleotide (1 mM) in 50 mM sodium cacodylate buffer, pH 7, in the presence of riboflavin (50 µM) for 2 minutes on a transilluminator (366 nm), under aerobic conditions at 4°C. Surprisingly for a photochemical reaction, the conversion is virtually quantitative. But the molecular gymnastics occurring are completely mind-blowing and we would invite reaction enthusiasts to review the proposed mechanism.
Product Information
8-OMe-dG-CE Phosphoramidite has been discontinued.
8-Amino-dA
8-Amino-2'-deoxyAdenosine has been substituted for 2'-deoxyAdenosine in oligonucleotides used in the studies of triple helix formation. It has been shown8 that oligonucleotides containing this modified base form stable triple helices at neutral pH, whereas regular triple helical structures are normally observed under acidic conditions. We are happy to make available 8-Amino-dA-CE Phosphoramidite (13), which can be used for oligonucleotide synthesis and subsequent deprotection without need for modification of normal procedures.
Product Information
8-Amino-dA-CE Phosphoramidite (10-1086)
UltraMild 2'-OMe-RNA
The use of UltraMild monomers in oligonucleotide synthesis has allowed very sensitive dyes like TAMRA, HEX and Cy5 to be used virtually routinely. The DNA and RNA monomers are currently available and, by popular demand, we are adding the set of 2'-OMe-RNA monomers. In our rendering of this chemistry, we use as protecting groups phenoxyacetyl (Pac) for A, acetyl (Ac) for C, and isopropyl-phenoxyacetyl (iPr-Pac) for G. The structures of the UltraMild 2'-OMe-RNA monomers are shown in Figure 1, (14) – (16).
It has become clear9 that acetic anhydride in the conventional capping mix can cause transamidation in situations where an amine protecting group is quite labile. This leads to acetyl protection on the amino group that may be slow to be removed. Consequently, if many dG residues are included in the oligonucleotide, we recommend the use of phenoxyacetic anhydride (Pac2O) in Cap A. This modification removes the possibility of exchange of the iPr-Pac protecting group on the dG with acetate from the acetic anhydride capping mix.



References
- (1) F. Seela and H. Driller, Helvetica Chimica Acta, 1988, 71, 1191-1198.
- (2) F. Seela and K. Kaiser, Helvetica Chimica Acta, 1988, 71, 1813-1823.
- (3) F. Seela and G. Becher, Nucleic Acids Res., 2001, 29, 2069-2078.
- (4) L.V. Nechev, I. Kozekov, C.M. Harris, and T.M. Harris, Chem Res Toxicol, 2001, 14, 1506-1512.
- (5) A.R. Diaz, R. Eritja, and R.G. Garcia, Nucleos Nucleot, 1997, 16, 2035-2051.
- (6) D.A. Erlanson, J.N.M. Glover, and G.L. Verdine, J. Amer. Chem. Soc., 1997, 119, 6927-6928.
- (7) H. Ikeda and I. Saito, J. Amer. Chem. Soc., 1999, 121, 10836-10837.
- (8) R.G. Garcia, E. Ferrer, M.J. Macias, R. Eritja, and M. Orozco, Nucleic Acids Res., 1999, 27, 1991-1998.
- (9) Q. Zhu, M.O. Delaney, and M.M. Greenberg, Bioorg Medicinal Chem Letter, 2001, 11, 1105-1107.
Product Information
- Glen Report 15.11: Redmond Red™ and Yakima Yellow™ dyes, and Eclipse™ Non-Fluorescent Quencher
- Glen Report 15.12: Comparison of Deprotection Methods for 3'-PT-Amino-Modifier CPG
- Glen Report 15.13: Pyrrolo-C - A Novel Fluorescent Nucleoside
- Glen Report 15.14: More and Still More Novel Minor Bases
- Glen Report 15.15: A Novel Route to Activated Carboxylate Modified Oligonucleotides











