Glen Report 29.15: Sequence Modification Using Glen Research's 5-Modified dU Family
Glen Research's first nucleoside for sequence modification, Amino-Modifier C6 dT (10-1039), was introduced in October 1989 and has led to a family of analogues that are useful for a variety of purposes. Although these are truly dU analogues, they behave as dT in primers and probes. By using the 5 position of the dU base (1), the attached tags project into the major groove of double stranded DNA where they are readily available for detection. Even extremely large molecules, like the enzyme horseradish peroxidase, can be attached at that position with minimal perturbation of normal hybridization.In this article, we will cover familial subsets designed for modification, fluorescent labelling, quenchers and click chemistry.
Modification
Amino-Modifier C6 dT (10-1039) has a long spacer designed to project the tag beyond the hybridization limit, whereas closely related Amino-Modifier C2 dT (10-1037) allows the tag to interact with adjacent DNA strands. In both cases, the primary amino group is revealed on deprotection, ready for post synthesis conjugation to a suitable tag, e.g., an N-hydroxysuccinimide (NHS) ester. In both cases, the amino group is protected as a trifluoroacetate (TFA).We have never found conditions which allow the TFA group to be removed from an amino-modifier while the oligonucleotide remains attached to the support. We are able to solve this problem by using a 9-fluorenylmethoxycarbonyl (Fmoc) protecting group. The Fmoc group can be removed on the synthesis column to allow on-column reaction with a variety of activated esters. Fmoc Amino-Modifier C6 dT (10-1536) is our dT analogue for this purpose.Carboxy-dT (10-1035) is hydrolyzed during deprotection with 0.4M methanolic sodium hydroxide (methanol:water 4:1) for 17 hours at room temperature and can be coupled directly to a molecule containing a primary amino group by a standard peptide coupling or via the intermediate NHS ester.With the introduction of NHS-Carboxy-dT (10-1535), it is possible to label one or multiple sites within an oligonucleotide with an amino tag while still on the support. This opens up the possibility to label any number of different dyes or molecules within an oligonucleotide when the phosphoramidite is unavailable. Doing so is straightforward and may be done manually off the synthesizer or even in a fully-automated manner on the DNA synthesizer. (http://www.glenresearch.com/Reports/gr23-16.html)Following many requests to allow a thiol to be created rather than an amino group, we introduced S-Bz-Thiol-Modifier C6-dT (10-1538) to join the ranks of thiol-modifiers for oligonucleotide synthesis. Thiol-Modifier C6-dT can be added as usual at the desired locations within a sequence.
Figure 1: Structures of Sequence Modifiers |
|
 |
 |
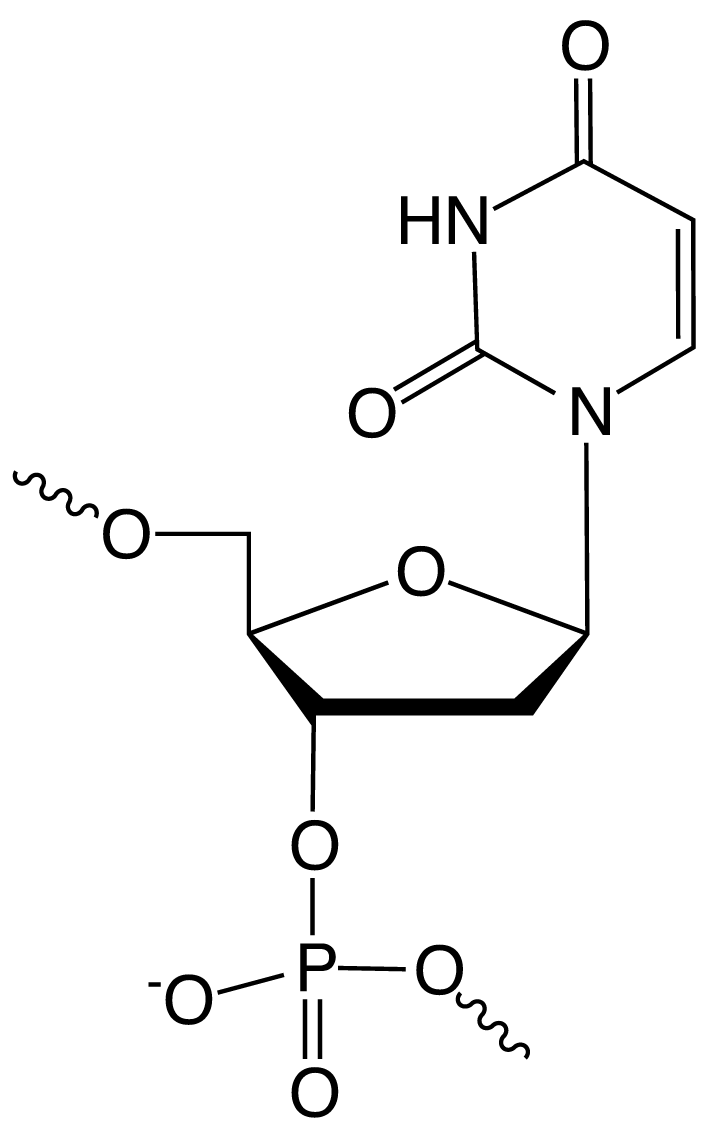
(1) 5 Position of dU |
10-1039 |
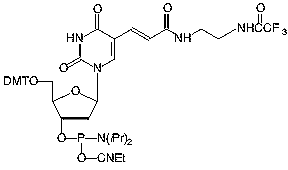 |
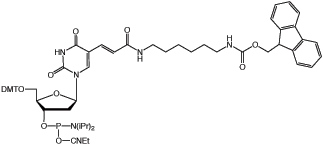 |
10-1037 |
10-1536 |
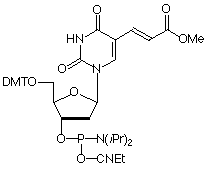 |
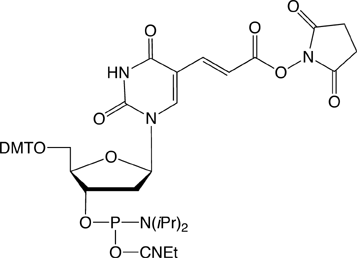 |
10-1035 |
10-1535 |
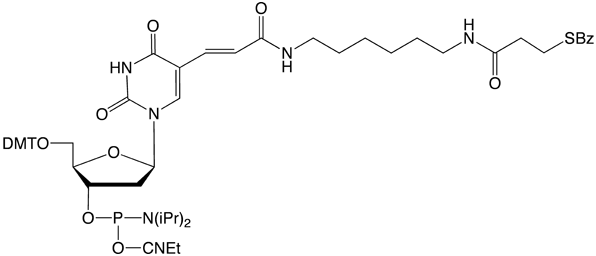 |
|
10-1538 |
|
LABELS
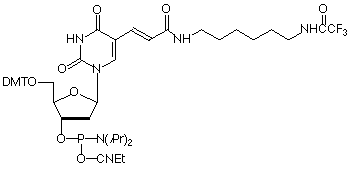
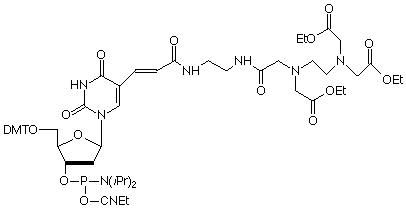
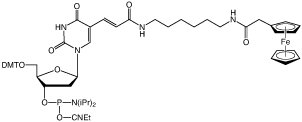
Of course, the ability to amino-modify oligos with Amino-Modifier C6 dT rapidly led to requests to have the most popular labels preinstalled. Biotin and fluorescein are the two most popular labels, so Biotin-dT (10-1038) and Fluorescein-dT (10-1056) were introduced.The earliest version of FRET probes used tetramethylrhodamine (TAMRA) as an internal quencher of the 5'-fluorophore. This was relatively inconveniently achieved using an oligo modified internally at a dT site with Amino-Modifier C6 dT and labelling post synthesis with TAMRA-NHS Ester. The introduction of TAMRA-dT (10-1057) allowed direct synthesis of such FRET probes.Molecular beacon probes and other hairpin probes have come to rely on the fluorescence quenching properties of the dabcyl molecule.A standard molecular beacon has a stem loop structure with a fluorophore like fluorescein at the 5'-terminus along with the quencher, usually a dabcyl group, at the 3'-terminus.However, in other hairpin probes, the fluorophore is again at the 5'-terminus but the dabcyl group is located within the sequence. The structure of the oligo should allow it to form a hairpin in such a way that the fluorophore is spatially adjacent to the quencher in the hairpin stem. The oligos can then be used as PCR primers. The fluorescence intensity of the amplified product correlates with the amount of incorporated primer since the hairpin no longer exists in the double-stranded amplified product. Dabcyl-dT (10-1058) can be used for direct incorporation into the quencher site.SIMA (HEX) is a more stable analogue of HEX which, unlike HEX, can be deprotected with AMA and SIMA (HEX)-dT(10-5945) is useful for sequence labelling.EDTA-C2-dT (10-1059) contains the triethyl ester of EDTA which allows sequence-specific cleavage of single- and double-stranded DNA and RNA. The cleavage reaction is only initiated once Fe(II) and dithiothreitol are added and so is readily controlled. Cleavage and deprotection of EDTA-C2-dT should be carried out with sodium hydroxide in aqueous methanol (0.4M NaOH in methanol/water 4:1) overnight at room temperature.With an excellent stability profile, ferrocene has always attracted considerable interest for DNA labelling to generate probes for electrochemical detection. Ferrocene-dT (10-1576) is easily added to oligonucleotides with no disruption of regular hybridization behavior.
|
|
 |
|
10-1038 |
|
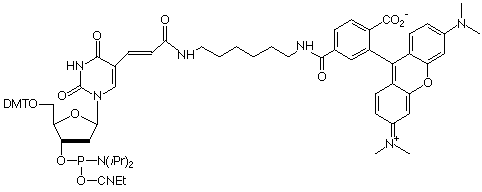 |
|
10-1057 |
|
 |
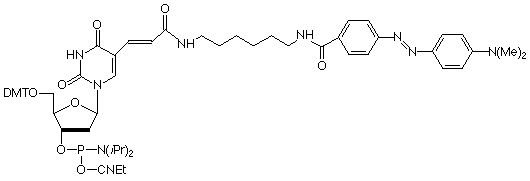 |
10-1056 |
10-1058 |
 |
|
10-5945; |
|
 |
 |
10-1059 |
10-1576 |
Quenchers
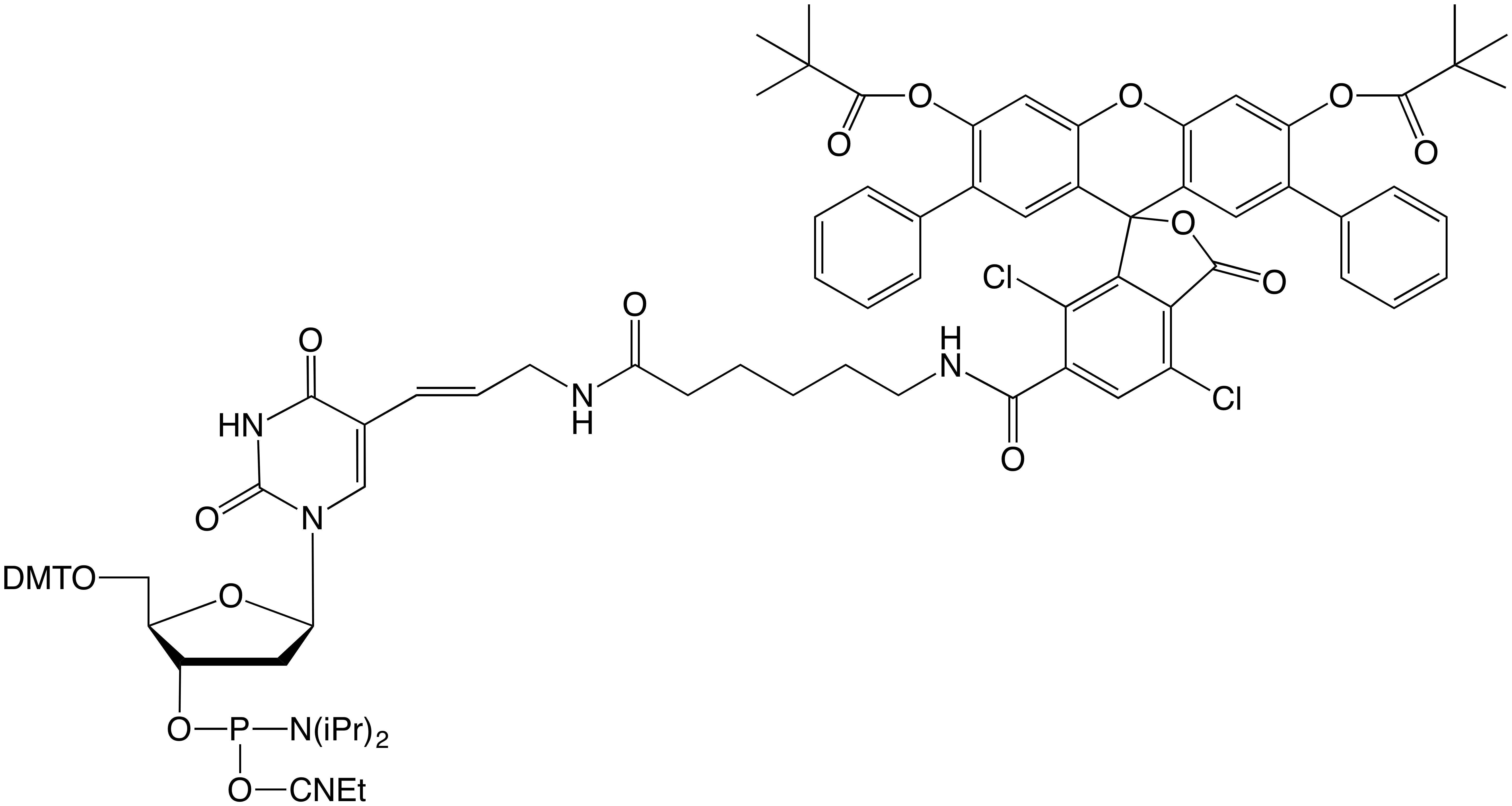
TAMRA and Dabcyl have both been used as quenchers of fluorescence by FRET or static quenching. However, other non fluorescent quenchers have become very popular over time. These include Black Hole Quenchers (BHQ), introduced by Biosearch Technologies, and Blackberry Quencher (BBQ), introduced by Berry & Associates.Two BHQ dyes are available from Glen Research as dT analogues, BHQ-1-dT (10-5941) and BHQ-2-dT (10-5942). These two BHQ dyes are suitable for quenching fluorescence in the range 450 - 650nm.BBQ-650®-dT (10-5944) has an absorption maximum of around 650nm and is capable of quenching fluorescence in the range 550 - 750nm.Figure 4 shows the wavelengths of fluorescence emission and quenching of various products available from Glen Research.
|
Figure 3: Structures of Quencher Modifiers |
|
 |
|
10-5941 |
|
 |
|
10-5942 |
|
 |
|
10-5944 |
|
Figure 4: Dye Fluorescence and Quencher Wavelength |
 |
Click Chemistry
The copper(I)-catalyzed azide-alkyne cycloaddition (CuAAC) reaction between azides and alkynes to form 1,2,3-triazoles was found to be exquisitely regioselective and efficient under very mild conditions. Although copper ions are known to damage DNA, typically yielding strand breaks, the use of copper stabilizing ligands in the reaction has allowed the CuAAC reaction to be used to functionalize alkyne-modified DNA nucleobases with extremely high efficiency. This strategy allows the use of the vast library of azide tags to be used in oligonucleotide labelling.
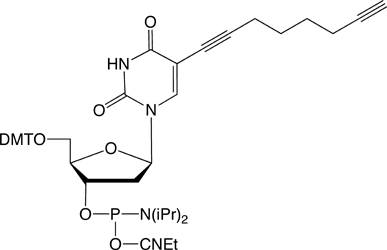
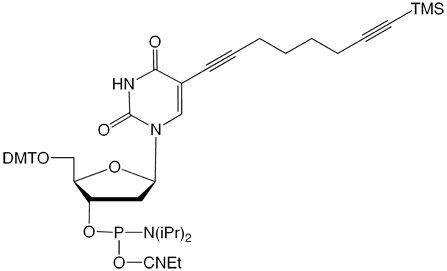
To realize efficient click-chemistry labelling of alkyne modified oligonucleotides, our dT analogues use a 5-(octa-1,7-diynyl) side chain. Oligonucleotides bearing a single nucleosidic alkyne group can be prepared using a C8-Alkyne-dT (10-1540). Using a combination of C8-Alkyne-dT, C8-TIPS-Alkyne-dT (10-1544) and C8-TMS-Alkyne (10-1545), it is possible to label oligonucleotides with different azide tags in up to three separate click reactions.
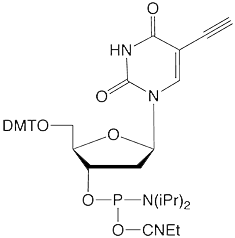
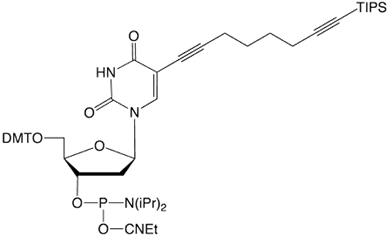
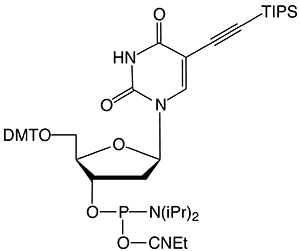
5-Ethynyl-dU (10-1554) offers convenient click conjugation with an azide to generate a label rigidly attached to one of the oligonucleotide bases. Mild deprotection conditions are necessary when using 5-Ethynyl-dU to prevent a hydration side reaction, which lowers the available alkyne level for the subsequent click reaction. TIPS-5-Ethynyl-dU (10-1555) contains an alkyne protected with a triisopropylsilyl (TIPS) group, which prevents acid or base catalyzed hydration during oligonucleotide synthesis and deprotection. A quick treatment with TBAF post synthesis removes the TIPS protecting group.
These click chemistry dT analogues are patent protected and available from Glen Research in collaboration with baseclick GmbH.
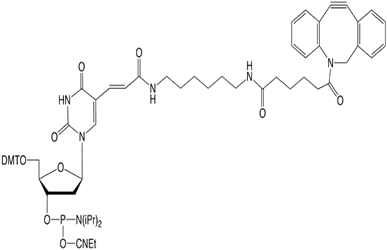
The dibenzocyclooctyl (DBCO) group exhibits the following desirable properties for use in oligonucleotide synthesis: simple to use; stable in solution on the synthesizer; stable to ammonium hydroxide and AMA; and excellent click performance in 17 hours or less at room temperature. DBCO-dT (10-1539) can be used for inserting a DBCO group at any position within the oligonucleotide and is then available for copper free click coupling with an azide.
Figure 5: Structures of Click Chemistry Sequence Modifiers |
 |
 |
10-1540 |
10-1545 |
 |
|
10-1544 |
10-1554 |
 |
 |
10-1555 |
10-1539 |
Product Information
Sequence Modifiers
Amino-Modifier C6 dT (10-1039)
Amino-Modifier C2 dT (10-1037)
Fmoc Amino-Modifier C6 dT (10-1536)
S-Bz-Thiol-Modifier C6-dT (10-1538)
Sequence Labels
Fluorescein-dT Phosphoramidite (10-1056)
SIMA (HEX)-dT Phosphoramidite (10-5945)
EDTA-C2-dT-CE Phosphoramidite (10-1059)
Ferrocene-dT-CE Phosphoramidite (10-1576)
Quencher Modifiers
BBQ-650®-dT-CE Phosphoramidite (10-5944)
Click Chemistry Sequence Modifiers
C8-Alkyne-dT-CE Phosphoramidite (10-1540)
C8-TMS-Alkyne-dT-CE Phosphoramidite (10-1545)-Discontinued
C8-TIPS-Alkyne-dT-CE Phosphoramidite (10-1544)-Discontinued
5-Ethynyl-dU-CE Phosphoramidite (10-1554)
- Glen Report 29.11: CDPI3 MGB™-Oligonucleotide Conjugates and Their Applications
- Glen Report 29.12: 5'-CDPI3 MGB™ Phosphoramidite and 3'-CDPI3 MGB™ CPG
- Glen Report 29.13: Studying DNA Repair Pathways Using Site-Specifically Modified Oligonucleotides
- Glen Report 29.14: N-Acetylgalactosamine (GalNAc) Oligonucleotide Conjugates
- Glen Report 29.15: Sequence Modification Using Glen Research's 5-Modified dU Family