Glen Report 20.14: More Click Chemistry
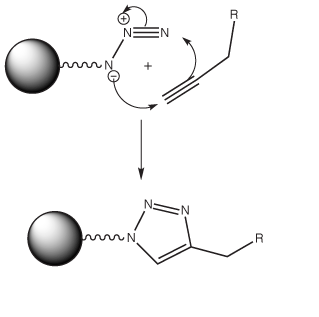
The term 'Click Chemistry' has been coined by Sharpless1 to describe the [3+2] cycloaddition2 between alkynes and azides, a reaction which has allowed remarkable selectivity in conjugation reactions in biological samples (Figure 1). Click chemistry is a very hot topic these days and click chemistry applied to oligonucleotides is developing rapidly with citations more than doubling each of the last two years. Without providing a definitive “Click” bibliography, here are some topics that have caught our interest recently: oligonucleotide cyclization with a microwave assist3; peptide-DNA conjugation4; oligonucleotide immobilization5, 6; fluorescent labelling7; oligonucleotides on gold nanoparticles8; and labeling following PCR with alkyne modified nucleoside triphosphates9.
|
|
|
|

|
In the Glen Report 19.1 (April 2007), we launched the 5’-Hexynyl Phosphoramidite (1) as an introduction to the application of click chemistry to oligonucleotides. This product has been well received but our offering was unbalanced in that the azide options were limited. However, we reported then that we were able to produce a 5’-azide by quantitatively converting a 5’-iodo modified oligonucleotide (prepared using 5’-Iodo-dT) or a 5’-dabcyl modified oligonucleotide (prepared using 5’-dabcyl-dT) using sodium azide. We are now happy to propose two new very simple ways for introducing an azido group into oligonucleotides.
For these new additions to our catalogue, we have been inspired by some publications and presentations from late 2007. In a presentation given during the 3rd meeting of the Oligonucleotide Therapeutics Society (Berlin, Germany, Oct 4-6, 2007), Tom Brown presented the work of his laboratory.10 He showed that very short cyclic oligonucleotides can be prepared in high yield on a multi-micromolar scale. They give rise to very stable duplexes that are potential tools for biophysical and biological studies. Cyclic oligonucleotides are very stable in serum for several days.
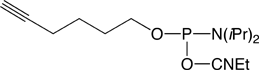
Almost at the same time, a French team at the Max Mousseron Institute for Biomolecules (Montpellier) developed the use of a bromohexyl phosphoramidite and different solid supports to allow conjugation and modifications.11
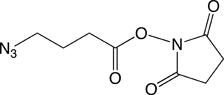
Our first proposal is to carry out the conjugation post-synthesis of an amino-modified oligonucleotide with an azide N-hydroxysuccinimide (NHS) ester, Azidobutyrate NHS Ester (2). This method is efficient for azido-modification of amines at either the 3’-end or the 5’-end of an oligo and it can even be used for internal modification on an Amino-Modifier-C6 dX residue within the sequence. The second approach, specific to the 5’-terminus, consists of adding 5'-Bromohexyl Phosphoramidite (3) in the last cycle. This modifier can then be easily transformed into a 5’-azido group by displacement of bromide using sodium azide. The first method can be performed in solution while the other allows the azide addition to be performed in solid phase on the synthesis column prior to cleavage and deprotection of the oligonucleotide. These products are easy to use and should prove valuable additions to our catalog.
Azidobutyrate Use
In this case, oligonucleotides are synthesized as usual on any synthesizer. Amino groups have to be introduced either at the 3’ or at the 5’ end of the oligonucleotide. After cleavage and deprotection, the azido group can be introduced as a modification of the amine. For example, the oligonucleotide in sodium carbonate/sodium bicarbonate buffer (pH 8.75) was incubated at room temperature with succinimidyl-4-azidobutyrate in DMSO. The final oligo is then purified using HPLC or precipitated with ethanol or butanol.
Bromohexyl Use
The 5'-Bromohexyl Phosphoramidite is used to make 5’-bromohexyl oligonucleotides with the same phosphoramidite elongation cycle used for the coupling as the regular 2'-deoxynucleoside phosphoramidites. Bromohexyl ODNs were then converted into azidohexyl ODNs on the column by treatment of the CPG with a solution of sodium azide and sodium iodide in dry DMF at 65 °C. Finally, the CPG with the 5’-azido oligo is treated with ammonia affording 5’-azidohexyl-ODNs in solution.11
Cyclization and Ligation of Oligos using Click Chemistry.
To exemplify the use and possibilities of this chemistry, we reference some work carried out in Professor Tom Brown’s laboratory at the University of Southampton (UK).10 This paper describes a template-directed oligonucleotide strand ligation, a covalent intramolecular DNA circularization and a catenation experiment using Click Chemistry. Tom Brown’s group also described the formation of a very stable cyclic DNA mini-duplex with just two base pairs, as an extension of this work.12
These experiments rely on an efficient and simple method of oligonucleotide click ligation that produces an unnatural extended DNA backbone linkage. One of the reacting ODNs contains a 3’-azide and the other a 5’-alkyne. The 5’-alkyne can be introduced using our 5’-Hexynyl Phosphoramidite (1).
For the oligonucleotide ligation reactions, the authors10 decided to use a strategy where a Cu[I] click catalyst was prepared in situ from Cu[II] sulfate and sodium ascorbate. All ligation reactions were carried out in 0.2 M aqueous sodium chloride to ensure complete formation of a duplex with the template. Unfortunately, under these conditions extensive degradation of all ODNs occurred, even with degassed buffers. However, the water-soluble tris-triazolylamine Cu[I]-binding ligand13 greatly reduced degradation and, when greater than 5-fold excess of ligand relative to Cu[I] was employed, very little decomposition was observed.
The ligated oligonucleotide product is obtained within two hours at room temperature using equimolar ratios of the two participating ODNs. The ligation can be forced to proceed with or without a template simply by varying the concentrations of the reactants. If the alkyne and azide are located in the same oligonucleotide, circularization occurs even in the absence of a template ODN. The circularized ODN can then be used in the formation of a double strand DNA catenane.
Examples with 12mer, 70mer and 72mer oligos were described.10 Click ligated single-strand cyclic oligos were purified by denaturing polyacrylamide gel electrophoresis. Circular ssDNA oligos were also prepared in this manner at 1.0 and 4.0 µmolar concentrations. Click ligation reactions of the same oligos were carried out at 10.0, 2.0, and 0.4 µmolar concentrations with and without a template oligonucleotide. Dilution was carried out by increasing the volume of 0.2 M aqueous NaCl. The authors observed that click chemistry can be conducted in a template-mediated manner over a wide concentration range, and below 2.0 µmolar the reaction does not proceed in the absence of a template ODN.
The authors analyzed the properties of the click ligation junction by conducting melting experiments in a quantitative PCR thermocycler (ROCHE Light-Cycler) in the presence of SYBR-Green.

(a) schematic and (b) chemical structure at ligation point.
(Illustration Courtesy of Reference 10)
Conclusion
Click chemistry is an excellent approach to carry out template-mediated ligation of two oligonucleotide strands against a complementary template. The methodology has also been shown to be efficient for the synthesis of a covalently closed ssDNA circle and a dsDNA pseudohexagon with sides of ca. 4 nm in length. This method is likely to be of value in nanotechnology applications involving DNA scaffolds. In addition, the use of single-stranded covalently closed DNA circles in biological applications warrants further investigation. Such constructs are likely to have increased in vivo stability, as they will be resistant to exonuclease degradation.
This click ligation may be useful in stabilizing structures like decoy oligos or aptamers. And such a fast and easy way of connecting two complementary oligo strands could be advantageous when compared to the synthesis of a single hairpin oligo of twice the length.
References
- V.V. Rostovtsev, Green, L.G., Fokin, V.V. and Sharpless, K.B., Angew Chem Int Ed, 2002, 41, 2596-2599.
- R. Huisgen, Angew Chem Int Ed, 1963, 2, 565-598.
- J. Lietard, A. Meyer, J.J. Vasseur, and F. Morvan, Journal of Organic Chemistry, 2008, 73, 191-200.
- K. Gogoi, M.V. Mane, S.S. Kunte, and V.A. Kumar, Nucleic Acids Res., 2007, 35, e139.
- M.V. Kvach, et al., Nucleosides Nucleotides & Nucleic Acids, 2007, 26, 809-813.
- D.I. Rozkiewicz, et al., Chembiochem, 2007, 8, 1997-2002.
- Y.H. Zhang, et al., Tetrahedron, 2007, 63, 6813-6821.
- M. Fischler, et al., Chem Commun (Camb), 2008, 169-71.
- P.M. Gramlich, C.T. Wirges, J. Gierlich, and T. Carell, Org Lett, 2008, 10, 249-51.
- R. Kumar, et al., Journal of the American Chemical Society, 2007, 129, 6859-6864.
- J. Lietard, A. Meyer, J.J. Vasseur, and F. Morvan, Tetrahedron Letters, 2007, 48, 8795-8798.
- A.H. El-Sagheer, et al., Chembiochem, 2008, 9, 50-2.
- T.R. Chan, R. Hilgraf, K.B. Sharpless, and V.V. Fokin, Org. Lett., 2004, 6, 2853-2855.
Product Information
- Glen Report 20.11: An Unnatural Base Pair System for the Expansion of Genetic Information
- Glen Report 20.12: Thiophosphoramidites and Their Use in Synthesizing Oligonucleotide Phosphorodithioate Linkages
- Glen Report 20.13: Technical Brief - Purification of 6-FAM Labelled oligos using Glen-Pak™ Cartridges
- Glen Report 20.14: More Click Chemistry
- Glen Report 20.15: Technical Brief - New application of 5-Me-iso-dC and iso-dG
- Glen Report 20.16: Technical Brief - New application for 5’-OMe-dT phosphoramidite
- Glen Report 20.17: New Product - Deuterated 2’-Deoxyguanosine Phosphoramidite
- Glen Report 20.18: New Products - Solid CPR II / Formylindole – Aldehyde Modifier
- Glen Report 20.19:New Product - 5'-Cholesteryl-TEG Phosphoramidite
- Glen Report 20.110: Improving Universal Support II for Oligonucleotide Synthesis
- Glen Report 20.111: Differences Between Universal Support II and III
- Glen Report 20.112: Technical Brief - Phage Display, Artificial Antibodies and Trimer Phosphoramidites

