Glen Report 17.13: UniCap Phosphoramidite, AN alternative to acetic anhydride capping
Introduction
The major impurities generated during oligonucleotide synthesis are n-1 and, to a lesser extent, n-2 deletion sequences. These deletion sequences are not homogeneous and result not from a single coupling failure but have been shown to be a statistical distribution of all possible n-1 or n-2 deletions by sequencing1, mass spectroscopy2 and hybridization to arrays of all possible complementary deletion strands.3 Deletion mutations can arise from an incomplete reaction at any of the steps during the synthesis cycle including; incomplete detritylation, incomplete capping of coupling failures or incomplete oxidation of the phosphite triester and subsequent hydrolysis back to the 5'-hydroxyl of the previous base during the next detritylation step. By far the predominant source of these deletions is incomplete capping of coupling failures during each synthesis cycle.
Standard capping is accomplished by acetylation of any remaining unreacted 5'-hydroxyls using a mix of acetic anhydride in THF (Cap A) and a capping activator, either dimethylaminopyridine (DMAP) or N-methylimidazole (MeIm) in THF (Cap B). A weak organic base, either pyridine or lutidine, is added to one of the Cap mixes.
The failure to cap, and the resulting generation of excess n-1 deletion sequences, present particular problems in trityl-on purifications and in the synthesis of long oligonucleotides for gene construction and cloning. Trityl-on purification relies on the increased hydrophobicity of the trityl group which is present only on the last base or monomer unit added and not on the capped failures. Unfortunately the n-1 deletions also possess 5'-trityls which make them elute along with the full-length oligo. These trityl-on deletions can be partially eliminated in HPLC purifications by collecting only the middle of the trityl-on peak since shorter deletion oligos elute on the backside of the peak. This is not possible with cartridge purification techniques, so final oligo purity is directly dependent on the ability to efficiently cap coupling failures. Long oligos are usually purified by denaturing PAGE and n-1 deletions represent the most difficult contaminant to remove, which explains why so many long oligos used for cloning are incorrect.
UniCap Phosphoramidite

In an attempt to improve overall synthesis fidelity, other approaches to capping have been explored. Since the coupling reaction is so efficient, one option is to use a phosphoramidite for capping. This option is the approach used in H-phosphonate chemistry. To that end the phosphoramidite of diethylene glycol monoethyl ether, UniCap, has been synthesized and compared to the standard capping mixes. Each capping mix was first evaluated for its ability to block oligo synthesis. Following a mock coupling using acetonitrile in place of amidite, three additional couplings were performed with the final trityl left on. This is an extreme case and represents a complete coupling failure. Quantification of the trityl-on peak represents the relative amount of capping failure. The results of these experiments conducted in quadruplicate are shown in Table 1.
The results clearly demonstrate that the capping is dependent on the activator in the Cap B solution. MeIm was a less effective catalyst for acetylation with 90% capping efficiency at 10% concentration. Increasing the concentration to 16% increases capping to 97%. UniCap performs substantially better at close to 99% capping, as seen in Table 1. Although DMAP is an extremely efficient catalyst for acetylation, its use has been reported to result in modification of O6-dG resulting in the formation of a fluorescent adduct.4 For this reason DMAP has been replaced in most Cap B mixes by MeIm.
| Synthesizer | Cap A Solution | Cap B Solution | Cap Efficiency (%) |
|---|---|---|---|
| Expedite | THF/Ac2O (9:1) | 10% MeIm/THF/Pyr (8:1) | 90.5 ± 1.9 |
| ABI | 394 THF/Pyr/Ac2O (8:1:1) | 10% MeIm/THF | 88.8 ± 2.5 |
| ABI | 394 THF/Lut/Ac2O (8:1:1) | 10% MeIm/THF | 89.1 ± 2.0 |
| ABI | 394 THF/Lut/Ac2O (8:1:1) | 16% MeIm/THF | 96.6 ± 1.4 |
| ABI | 394 THF/Lut/Ac2O (8:1:1) | 6.5% DMAP/THF | 99.4 ± 0.3 |
| ALL | UniCap Phosphoramidite | 98.6 ±0.4 |
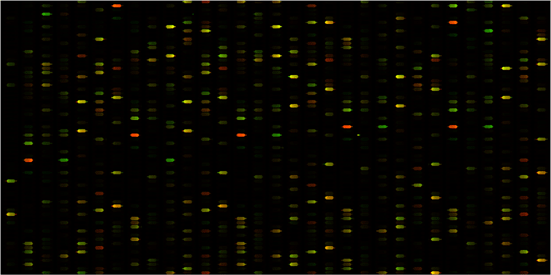
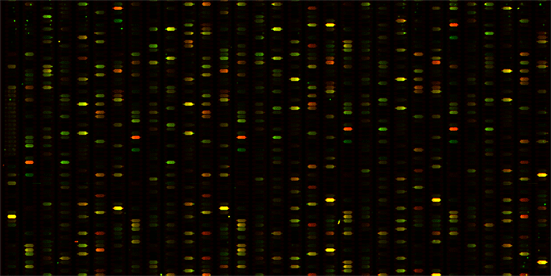
Cy3(green): universal mRNA_cDNA
Cy5 (red): skeletal muscle mRNA_cDNA
Capping with UniCap Phosphoramidite
To use UniCap as a capping amidite on the Expedite 8909 or ABI synthesizers, dilute it to the standard amidite concentration and place the vial in position 5 on the instrument. ABI cycles can be modified by adding coupling steps after the last column coupling step "Column Off", replacing "Base + Tetrazole to Column" (Function 33) with "5 + Tetrazole to Column" (Function 35). For use on the Expedite synthesizer, copy the coupling steps for amidite reservoir 5 and paste them into the coupling section of each of the other amidite cycles. The standard capping steps can be left out of the cycle. Although we have been unable to confirm that acetate capping reverses any O6-dG modification formed during coupling, regular capping can certainly be left in the cycle if this is a concern.
When UniCap Phosphoramidite was tested, it was found to be very highly effective at nearly 99% capping efficiency and, in addition, was determined to be stable for at least one week on the synthesizer.
UniCap Phosphoramidite was originally developed for capping in oligo synthesis on the surface of chips. Capping is often omitted in this situation because acetylation by acetic anhydride can change the polarity and surface characteristics of the chips. UniCap provides virtually quantitative capping without changing the polarity of chip surfaces. This reduces the background and increases the contrast of the array fluorescence. We are grateful to Dr. Xiaolian Gao, University of Houston, for providing the following information.
Oligonucleotide Microarray Synthesis on Microfluidic Chip
An oligonucleotide microarray containing 3888 sequences, which are selected from human cancer related genes, was synthesized as described previously.5 One chip synthesis used a regular protocol with acetic anhydride (AC) capping and the other chip used the same protocol except for UniCap Phosphoramidite (PEG) capping.
DNA Chip Hybridization Using cDNA Samples
Two cDNA samples were prepared according to procedures described (). The universal (univ) and skeletal muscle (sk) total RNA was from Clontech. Fluorescent Cy3 and Cy5 dyes were incorporated using dye-dU for the univ and sk cDNA samples, respectively. The co-hybridization of the cDNA samples to the DNA chip used 6' SSPE (0.9 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) buffer (80 mL) mixed with 25% formamide at 32 °C for 18 h under micro-flow conditions. The chips were washed briefly with the 6' SSPE buffer before image scanning on an Axon GenePix 4000B laser scanner. The PMT level was adjusted according to the signal strength observed. The images of the AC and PEG capping DNA chips are shown in Figure 2.
Validating Oligonucleotide Synthesis on a Chip Using Hybridization
The PEG capping was implemented in DNA chip synthesis and the comparison chip was synthesized using regular AC capping. The goals in these experiments were to compare hybridization results when the two DNA chips were treated with cDNA samples labeled with Cy3 (universal total RNA sample) or Cy5 (skeletal muscle total RNA sample) fluorescent dye. The two samples were co-hybridized to the chip and the ratio of Cy3 to Cy5 is shown in color ranging from green (Cy3 > Cy5) to yellow (Cy3 = Cy5) to red (Cy5 > Cy3). The color ratio image comparison of the PEG capping versus the AC capping chip is shown in Figure 2. This experiment validates that PEG capping is applicable to DNA microarray synthesis for the improvement of capping efficiency, which is critical for the initial synthesis steps of in situ oligonucleotide synthesis on glass surfaces. In addition, the capping reaction time using the PEG capping reagent is several fold shorter than that of AC capping.
The capping reagent was developed by Peilin Yu and Xiaolian Gao, Department of Chemistry, University of Houston. UniCap Phosphoramidite is sold by Glen Research under license from the University of Houston and is intended for research purposes only.
References
- J. Temsamani, M. Kubert, and S. Agrawal, Nucleic Acids Res., 1995, 23, 1841-1844.
- K.L. Fearon, J.T. Stults, B.J. Bergot, L.M. Christensen, and A.M. Raible, Nucleic Acids Res., 1995, 23, 2754-2761.
- D.H. Chen, Z.M. Yan, D.L. Cole, and G.S. Srivatsa, Nucleic Acids Res., 1999, 27, 389-395.
- J.S. Eadie and D.S. Davidson, Nucleic Acids Res., 1987, 15, 8333-49.
- X.L. Gao, et al., Nucleic Acids Res., 2001, 29, 4744-4750.
Product Information
- Glen Report 17.11: Introduction
- Glen Report 17.12: Products for siRNA Research
- Glen Report 17.13: UniCap Phosphoramidite, AN alternative to acetic anhydride capping
- Glen Report 17.14: Expanding Our Repertoire of Dark Quenchers: Black Hole Quenchers
- Glen Report 17.15: 2'-Fluoro-RNA Monomers
- Glen Report 17.16: Internally Quenched Nucleotide Fluorescent Reporters
- Glen Report 17.17: AB 3900 and MerMade Columns

