Glen Report 13.12: A Rapid Method for Atomic Mutagenesis of Nucleic Acids
Interference Mapping
Nucleotide Analog Interference Mapping (NAIM) is a powerful new approach to perform atomic mutagenesis simultaneously, yet individually, at every position in an RNA using materials easily prepared by in vitro transcription.1 Through the use of a collection of nucleotide analogs, now available from Glen Research, it is possible to assess the importance of every 2'-OH, exocyclic amine, or imino group throughout the length of an RNA. By comparing the results of multiple nucleotide analogs, it is possible to determine which ribose rings adopt unusual sugar puckers, which 2'-OH groups serve as hydrogen bond donors and/or acceptors, which nucleotides have perturbed pKas for potential use in folding or catalysis. Furthermore, this approach can reveal which nucleotides make tertiary contacts, on which nucleotide face the contacts are made, and where metal ions are coordinated within an RNA fold.
This extraordinary level of chemical detail is available quite rapidly using a collection of nucleotide analogs that modify individual functional groups on the RNA base or ribose sugar. The analogs are all a-thiotriphosphates which mark the sites of their incorporation within the transcript with an a-phosphorothioate linkage2. This linkage allows simple iodine cleavage and makes it possible to identify the sites where incorporation of the analog interfered with or enhanced the activity of the RNA molecule. The magnitude of an interference effect at each position in the molecule can be quantified by simply viewing a sequencing gel of the reaction products (Figure 1A).
A complete description of this approach was the subject of Glen Report, 1998, 11(2), 6-8, and can be accessed at: GR11-24.html
An up-to-date listing of the available a-thiotriphosphates is maintained at: The a-thiotriphosphates have been discontinued.
This rapid and inexpensive alternative to single site atomic mutagenesis has been implemented for the study of several systems, including RNA folding,3 ribozyme catalysis,4-6 RNA-protein interactions,7,8 and even in vivo RNA modification. The method can be applied to the study of any nucleic acid that can be functionally selected from among a larger pool of less active variants. Selection schemes have included native gel mobility shifts, nitrocellulose filter binding, selective radiolabeling by RNA ligation, and size selection in a denaturing polyacrylamide gel.3-8 It has been applied toward the study of catalytic RNA reactivity, tRNA-synthetase interactions, mRNA processing, rRNA modification and snoRNA assembly. RNAs successfully studied by NAIM range from less than 50 to almost 1000 nucleotides in size, yet in all cases the data defines the contribution of a specific functional group at the individual nucleotide level.
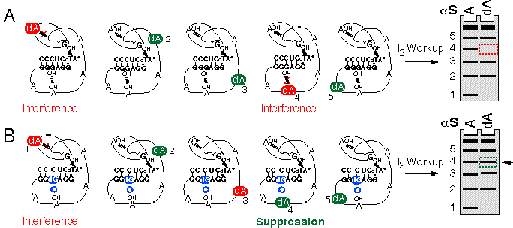
In this particular example, the RNA has five A's and the 2'-OH groups of two of them (1 and 4 shown in red) are important for activity based upon the NAIM experiment with dAaS (A). Interference suppression is performed in the same manner as NAIM, but in this case a 2'-OH known to be important for activity is substituted with a 2'-deoxy G nucleotide within all the molecules in the population. This eliminates one of the critical hydrogen bonds (blue circle) and causes all of the molecules in the population to be less active. However, deletion of the second hydrogen bond in the pair has no additional deleterious effect on activity, so it can be removed without significant penalty. In this example, specific suppression of dAaS interference is observed at A4, which is evidence that the 2'-OH of A4 and the 2'-OH of the substituted G participate in a hydrogen-bonding interaction.
Interference Suppression
Although NAIM provides a great deal of chemical information about the functional importance of each nucleotide in an RNA sequence, it can be difficult to reach specific conclusions about the tertiary structure from primary interference data alone. While a single NAIM experiment can identify all of the important 2'-OH groups and exocyclic amines within an RNA, it does not define how these groups interact within the overall RNA fold. However, such information can be obtained using a variation of NAIM, termed Interference Suppression.9 This method, which combines site-specific analog substitution and interference modification approaches, makes it possible to identify specific tertiary hydrogen-bonding partners within an RNA structure.10 The principle behind the approach is that, if an interaction is disrupted by deletion or alteration of one functional group in an interacting pair, then no additional energetic penalty will result from deletion or alteration of the second functional group in the pair. Interference suppression is scored by the reappearance of a specific band on the sequencing gel which showed interference in the context of the initial NAIM experiment (Figure 1B).
For example, if every RNA in the population lacks a 2'-OH group that is known to be important for activity, interference will be suppressed at the specific site with the specific functional group that makes a hydrogen bonding interaction with the deleted group. This type of experiment has provided a series of hydrogen bonding structural restraints that have made it possible to construct a detailed model of the group I intron active site (Figure 2).10 Within this catalytic RNA, deletion of the 2'-OH at G22, resulted in suppression of 2'-deoxy and 3-deaza interference at A114 (red atoms in Figure 2), while similar suppressions were observed exclusively at A207 if the exocyclic amine of G22 was deleted (yellow atoms in Figure 2).9 Suppression experiments do not necessarily require single atom substitution, as mutation of a specific amino acid residue in a bound protein or mutation of a nucleotide base involved in a tertiary contact can also accomplish a similar effect.
Nucleic acid biochemistry is about to enter a new era. With the soon to be revealed high resolution structure of the ribosome, the vast majority of the structural motifs available to RNA will be known, as will many of the rules by which proteins interact with RNA molecules. The next challenge will be to identify high resolution biochemical signatures for these folding motifs and to assign biological function to the amazingly complex structures that nucleic acids are capable of adopting. NAIM provides a particularly valuable tool in this effort and Glen Research is pleased to make this collection of reagents available to the scientific community for the multitude of applications that can be envisioned for them.
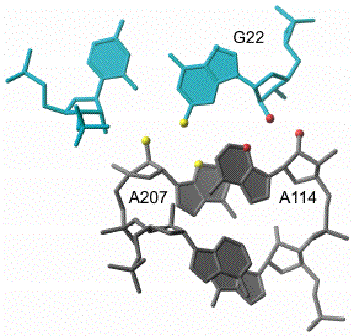
The sets of interacting functional groups identified by interference suppression are shown in yellow and red spheres of larger radius. The sheared A·A pairs in the J4/5 segment of the active site are shown in gray and the G·U wobble pair in the P1 helix is shown in cyan. The nucleotide numbers within the Tetrahymena group I intron are also indicated. See text for discussion of specific interactions.
References
- S. A. Strobel, Curr Opin Struct Biol, 1999, 9, 346-352.
- G. Gish and F. Eckstein, Science, 1988, 240, 1520-1522.
- S. Basu, R. P. Rambo, J. Strauss-Soukup, J. H. Cate, A. R. Ferre-D Amare, S. A. Strobel and J. A. Doudna, Nat. Struct. Biol., 1998, 5, 986-992.
- S. A. Strobel and K. Shetty, Proc. Natl. Acad. Sci. U.S.A., 1997, 94, 2903-2908.
- M. Boudvillain and A. M. Pyle, EMBO J., 1998, 17, 7091-7104.
- A. V. Kazanstev and N. R. Pace, RNA, 1998, 4, 937-947.
- R. T. Batey, R. P. Rambo, L. Lucast, B. Rha and J. A. Doudna, Science, 2000, 287, 1232-1239.
- C. S. Vortler, O. Fedorova, T. Persson, U. Kutzke and F. Eckstein, RNA, 1998, 4, 1444-1454.
- S. A. Strobel, L. Ortoleva-Donnelly, S. P. Ryder, J. H. Cate and E. Moncoeur, Nat. Struct. Biol., 1998, 5, 60-66.
- S. A. Strobel, Biopolymers, 1998, 48, 65-81.
- Glen Report 13.11: C-5 Propynes, Cytofectin and Other Product Updates
- Glen Report 13.12: A Rapid Method for Atomic Mutagenesis of Nucleic Acids
- Glen Report 13.13: Trityl Group in the 3rd Millenium: New Perspectives for Oligonucleotide Chemistry and Beyond
- Glen Report 13.14: Polyplex Synthesizer from Genemachines
- Glen Report 13.15: New Monomers: 2,4-Difluorotoluene, Fluorescein Phosphoramidite and Support, 2',3'-Dideoxynucleosides
- Glen Report 13.16: Product Update - Which 5'-Amino-Modifier?

