Glen Report 28.21: Versatile Applications of the Copper(I)-Catalyzed Click Chemistry
Authors: Birgit Oberleitner1, Sascha Serdjukow1, Thomas Carell2 and Thomas Frischmuth1
Address: 1 baseclick GmbH, Floriansbogen 2-4, 82061 Neuried, Germany, email: info@baseclick.eu
2 Ludwig-Maximilians-Universität (LMU) München, Butenandtstr. 5-13, 81377 Munich, Germany
Note: Products of baseclick are patent protected and available from Glen Research Corporation, 22825 Davis Drive, Sterling, VA 20164, USA, email: support@glenresearch.com, in collaboration with baseclick.
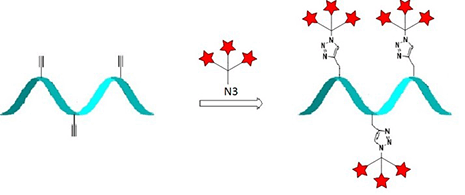
Fluorophore azide clicked to EdU
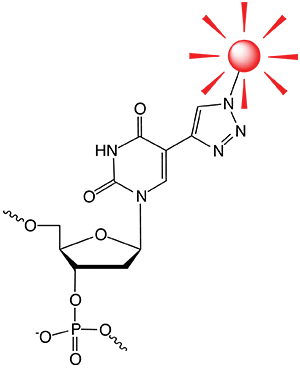
Introduction
Since the discovery of the copper(I)-catalyzed reaction between azide and alkyne (CuAAC)-moiety in 2002,[1] its unique characteristics, high yield and mild reaction conditions, have made it one of the most prominent if not “the” click reaction. Although the purpose of this reaction, joining two molecules, is very simple, the plethora of applications that make use of this efficient chemistry demonstrates that only creativity and imagination may be the main limits for its application. Using CuAAC for the modification of oligonucleotides has been particularly fruitful [2] and is the foundation of the company baseclick. The following applications exemplify the broad scope of the method.
Applications
1) Click Chemistry for Cell Proliferation Detection
The detection of cell proliferation is of utmost importance for cell imaging, the diagnosis of genotoxicity, the evaluation of anti-cancer drugs, to name just a few. In order to detect DNA Synthesis of dividing cells, an alkyne bearing thymidine analogue, EdU (5-Ethynyl-2'-deoxyuridine), is incorporated into DNA during cell division.[3] For analysis, the fluorophore azide is subsequently “clicked” to the sample and detected using various methods (see Figure 1, Page 2). Only cells which have newly synthesized DNA are detected by microscopy, fluorescence-activated cell sorting (FACS) techniques or high throughput screening (HTS). Notably, the EdU method has high selectivity, sensitivity and reproducibility. Moreover, it allows nearly unrestricted choice of the fluorescent label.
Currently, a combined approach by baseclick and the group of Prof. Thomas Carell (LMU Munich) aims for further improved sensitivity of the technology.
Figure 1: Schematic illustration of the click chemistry based EdU cell proliferation assay
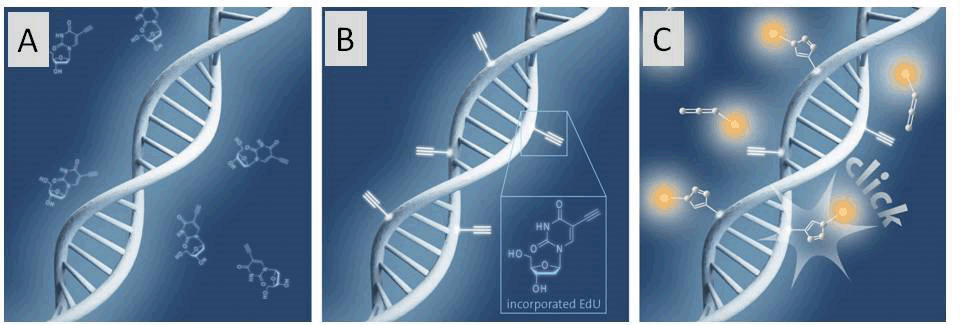
A) Incubation of cells with EdU. B) EdU is incorporated during active DNA synthesis. C) Detection of proliferating cells via click chemistry, whereby a variety of different fluorescent dye azides can be used.
Figure 2: Schematic representation of a DNA tube
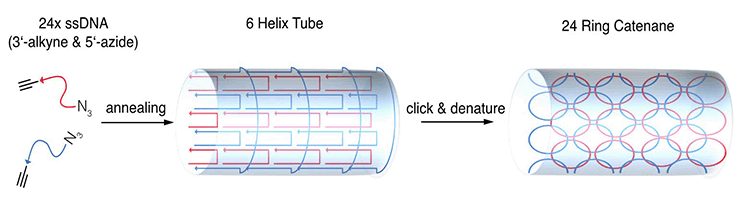
Schematic representation of a DNA tube consisting of 24 (3’-alkyne, 5’-azide)-modified oligonucleotides. The pre-organized position of the click reactive groups within the tube is achieved by strand hybridization. After addition of a click catalyst, the intramolecular reaction is carried out yielding a DNA catenane.
Figure 3: Left: FISH assay and Right: Current strategy for signal enhancement
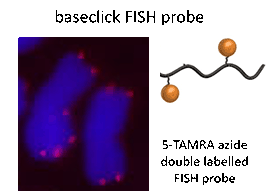
Left: FISH assay (red probe) on mitotic chromosomes of rye (Secale cereale) using two labels per FISH probe. Right: Current strategy for signal enhancement using dendritic structures.
2) DNA nanotechnology
DNA nanotechnology is an emerging research field. The user-defined self-assembly of nucleic acids to construct DNA nanostructures of various shapes opens new DNA applications[4], e.g., as drug delivery tools, a platform for single molecule reaction, molecular machines and many more. Unfortunately, these complex structures require high salt concentrations to maintain correct folding.
By applying click chemistry to DNA nanotechnology, baseclick has created very stable nanostructures.[5] A DNA nanotube (24 nm in length) was designed from a set of doubly modified oligonucleotides (alkyne and azide functionality on each oligonucleotide). After folding of the nanotube, the strands were interlocked at the ends by CuAAC (Figure 2). The nanotube is so robust that it remains intact even under denaturing conditions and in the presence of nucleases.
3) Next Generation DNA-FISH and mRNA FISH Probes
Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that enables the detection and localization of specific DNA or RNA sequences on chromosomes and cell compartments (target). The principle of this method is to hybridize labelled single stranded nucleic acid molecules such as DNA and RNA (probe) to complementary target sequences.
Baseclick FISH probes are oligonucleotides containing two or three reporter molecules attached by CuAAC for improved signal intensity.[6] These FISH probes are hybridized with the target and detected subsequently by microscopy. A strategy where clickable FISH probes are hybridized with the target and labelled afterwards is also feasible. This so-called post-hybridization click condition allows the use of sensitive reporter molecules that are mostly not available to standard FISH probes. Baseclick FISH probes display a high signal-to-noise ratio, possess high labelling rates and have an easy-to-handle protocol (Figure 3).
At the moment, a combined effort of baseclick and the Carell group at the LMU Munich is aiming for further improved sensitivity of the method using dendrimer structures for increased fluorescence of the probe.
4) CuAAC for Preparation of Bispecific Aptamers
Aptamers are oligonucleotide sequences (DNA or RNA) which have outstanding potential for applications in biosensing, bioimaging and cancer therapy. They specifically bind another molecule (e.g., nucleic acids, proteins, small organic compounds) in a manner similar to antibodies. Aptamers can be prepared by solid-phase synthesis. Thus they are commercially available and can be easily modified site-specifically in contrast to antibodies. Using the CuAAC technology, baseclick has prepared aptamers with two different binding specificities combined in a single aptamer molecule by efficiently joining two aptamers. These aptamers have similar applications as bi-specific antibodies and thus could bring together two different cells (e.g., cancer cell and T cell) and induce targeted cancer cell destruction.
Conclusion
Other applications of the CuAAC that have been developed within the group of Prof. Carell in recent years include synthesis of labelled nucleotides,[7] and synthesis of probes for the sequencing of epigenetic markers.[8] Despite the numerous applications which have already been developed using CuAAC, many more are expected in the near future. Using new ligands and applying improved reaction conditions, researchers have clicked on proteins,[9] on glycan structures and even in vivo in fish embryos.[10]
References
[1] a) V. V. Rostovtsev, L. G. Green, V. V. Fokin, K. B. Sharpless, Angew. Chem. Int. Ed. 2002, 41, 2596-2599. b) C. W. Tornoe, C. Christensen, M. Meldal, J. Org. Chem. 2002, 67, 3057–3064.
[2] A. H. El-Sagheer, T. Brown, Chem. Soc. Rev. 2010, 39, 1388–1405.
[3] a) A. Salic, T. J. Mitchison T J, Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. b) B. Oberleitner, A. Manetto, T. Frischmuth, Biospektrum 2014, 02, 188. c) S. Diermeier-Daucher, S. T. Clarke, D. Hill, A. Vollmann-Zwerenz, J. A. Bradford, G. Brockhoff, Cytometry A 2009, 75, 535–546.
[4] D. S. Lee, H. Qian, C.Y. Tay, D.T. Leong, Chem. Soc. Rev. 2016, 45, 4199–4225.
[5] V. Cassinelli, B. Oberleitner, J. Sobotta, P. Nickels, G. Grossi, S. Kempter, T. Frischmuth, T. Liedl, A. Manetto, Angew. Chem. Int. Ed. 2015, 54, 7795–7798.
[6] a) A.M.Oliveira, C.A. French, Acta Cytol. 2005, 49,587–594. b) E.V. Volpi, J.M. Bridger, BioTechniques 2008, 45, 385–409.
[7] S. Serdjukow, F. Kink, B. Steigenberger, M. Tomas-Gamasa, T. Carell, Chem. Commun. 2014, 50, 1861–1863.
[8] M. Su, A. Kirchner, S. Stazzoni, M. Müller, M. Wagner, A. S. Schröder, T. Carell, Angew. Chem. Int. Ed. 2016, 55, 11797–11800.
[9] C. Uttamapinant, A. Tangpeerachaikul, S. Grecian, S. Clarke, U. Singh, P. Slade, K. R. Gee, A. Y. Ting, Angew. Chem. Int. Ed. 2012, 51, 5852–5856.
[10] C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu, P. Wu, Angew. Chem. Int. Ed. 2011, 50, 8051–8056.
Intellectual property rights:
baseclick GmbH has been granted the following patents (1-3) besides its further patent applications (4-5).
1. WO 2006/117161 (New labelling strategies for the sensitive detection of analytes)
2. WO 2008/952775 (Click chemistry for the production of reporter molecules)
3. WO 2010/115957 (Click Chemistry on heterogeneous catalysts)
4. PCT/EP 2013/064610 (Anandamide-modified nucleic molecules)
5. PCT/EP 2015/056007 (Self-assembly of DNA Origami: a diagnostic tool)
baseclick GmbH holds a worldwide exclusive license for granted patent application
WO 03/101972 (Copper-catalysed ligation of azides and acetylenes for the nucleic acid field) in the area of diagnostics and research. As Glen Research and baseclick are partners, Glen Research is now able to help in sublicensing this outstanding technology.
Product Information
5-Ethynyl-dU-CE Phosphoramidite (10-1554)
- Glen Report 28.21: Versatile Applications of the Copper(I)-Catalyzed Click Chemistry
- Glen Report 28.22: New Product - TIPS-5-Ethynyl-dU-CE Phosphoramidite
- Glen Report 28.23: Technical Brief – Fluorescein Labelling – Considering the Options
- Glen Report 28.24: New Products – AquaPhluor® 593 5’-Phosphoramidite and CPG
- Glen Report 28.25: New Product: Glen-Pak™ DNA 3mg 384-Well Plate
- Glen Report 28.26: Transient Cyanoethylation - An Unexpected Route to Chemical Bleaching of a Fluorophore

