Glen Report 11.15: Update - Universal Supports, Q-supports
Universal Support
Until our recent addition of a Universal Support, procedures in oligonucleotide synthesis required that the solid support contain the first nucleoside. This situation therefore required that an inventory of all four regular nucleoside supports be maintained. At the same time, oligonucleotides with unusual nucleosides, available as phosphoramidites but not as supports, at the 3'-terminus could not be readily prepared. However, the most worriesome aspect of this situation is the potential for a mistake to be made in the selection of the column containing the 3'-nucleoside. This potential for error may be fairly low in regular column-type synthesizers, but it is especially significant in the new generation of parallel synthesizers where 96 or 192 wells may contain all four supports in a defined grid.
Our initial choice as Universal Support(1), shown below, allows the detritylation of the support to be carried out under normal conditions, as is the addition of the first nucleoside monomer. The remainder of the oligonucleotide synthesis also proceeds without any changes from the regular cycles.

The base-mediated elimination of the terminal phosphodiester group in the oligonucleotide - the mechanism is shown in Figure 2 - is the crucial step in this strategy. The elimination must proceed promptly under conditions comparable with routine deprotection strategies, using reagents which are also standard. Preferably the reagents should be volatile for simple evaporation (ammonium hydroxide or aqueous methylamine) or readily desalted (aqueous sodium hydroxide).
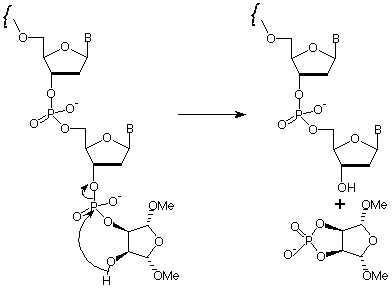
Optimization of conditions for the elimination of this structure to give 3'-OH at the terminus has led to a selection of conditions which have been readily adopted in many labs (Table 1). Using ammonium hydroxide, oligonucleotide deprotection and elimination to the 3'-OH is carried out at 80°C for a minimum of 8 hours, but preferably overnight. As long as acetyl protected dC monomer (Ac-dC) is being used, deprotection and elimination can be carried out in AMA at 55°C overnight, or at 80°C for a minimum of 3 hours. Both of these cleavage and elimination solvents are completely volatile and standard procedures can be applied for desalting and/or purification.
Again as long as Ac-dC monomer is being used, very fast deprotection and elimination occurs in aqueous alcoholic sodium hydroxide at 80°C for 30 minutes. Since this solution contains no volatile gases, very little pressure build-up occurs at 80°C. This mixture can be diluted with water and desalted using standard techniques.
Alternatively, 0.2 µmole or greater syntheses can be ethanol precipitated and the desalted, partially-purified sodium salt of the oligo can be quickly isolated by centrifugation.
Using the Universal-Q support, the oligonucleotide can be cleaved from the support in a matter of a few minutes using any of the above deprotection and elimination solutions. The elimination reaction must then be continued for the indicated time. The Universal Support (1) is sold under license from Zeneca PLC.
Product Information
Q-Supports
Cleavage of the succinate linkage between the the oligo and the support using ammonium hydroxide, either manually or on the synthesizer, is a major bottleneck in productivity for many synthesis groups. It consumes one hour of precious time while releasing only about 80% of the oligonucleotide. Richard Pon's group has identified2 hydroquinone-O,O'-diacetic acid as the most satisfactory alternative to the succinate group. Nucleosides with this linker arm (Q-linker) are attached to supports with the same ease as the succinyl linker arm. The cleavage time in ammonium hydroxide at room temperature was found to be a mere 2 minutes while the linkage was shown to be very stable to the reagents of routine oligonucleotide synthesis.
The Q-linker is absolutely compatible with all hydrolytic cleavage procedures, especially mild procedures like potassium carbonate in methanol. Glen Research has been offering Q-supports under license from the University of Calgary for around 6 months. Initially, we have made available Q-linkers on 500Å CPG in 0.2 and 1 µmole scales in both Applied Biosystems and Biosearch formats with the four most popular nucleosides being offered, as well as the Universal Support.
The response to date has been quite gratifying. Interestingly, the main usage of the Q-supports so far has been in Biosearch columns where cleavage has normally been carried out manually. Clearly, reducing the manual step from 1 hour to 2 minutes hUniversal Support IIIas been the critical selling point for Biosearch customers. We look forward to increased use of Q-supports to increase throughput on the popular Expedite instruments with 16 column add-ons.
Although in use on Applied Biosystems' instruments by several groups, the lack of columns in the low volume format is clearly impeding adoption of the Q-supports. We are actively working on a low volume Q-support column for LV40 and LV200 cycles and expect columns to be available in the fourth quarter of this year.
References
S. Scott, P. Hardy, R.C. Sheppard, and M.J. McLean, Innovations and Perspectives in Solid Phase Synthesis, 3rd International Symposium, 1994, Ed. Roger Epton, Mayflower Worldwide, 115-124.
R.T. Pon and S.Y. Yu, Tetrahedron Lett., 1997, 38, 3327-3330.
Product Information
- Glen Report 11.11: Using Modified Bases to Optimize Hybridization
- Glen Report 11.12: Oligo Affinity Supports
- Glen Report 11.13: New Fluorescent Reagents - TAMRA-dT, Dabcyl-dT
- Glen Report 11.14: The Use of Puromycin CPG in Combinatorial Chemistry
- Glen Report 11.15: Update - Universal Supports, Q-supports
- Glen Report 11.16: More Novel Monomers - 6-Thio-dG, 2'-Amino-RNA, 5-Hydroxy-dC,dU Spacer C18, Spacer 9-CPG

