Glen Report 36-26: Technical Note Preventing Detritylation During RNA Deprotection
In our last issue, we reported on coupling efficiency using various monomers (DNA vs. TOM vs. TBDMS).1
As expected, TOM performed better than TBDMS, which has a more significant impact for longer oligonucleotides. During our experiments, we observed some unavoidable loss of the DMT group while drying the oligonucleotide down between global deprotection/cleavage and 2′-desilylation. This is not ideal, especially for those relying on DMT-ON purification for their RNAs. To follow up on this, we sought a method to minimize detritylation during the workup of RNA.
It turns out, we didn’t have to look too far. About 15 years ago, we developed a method to prevent detritylation caused by drying down and tested this on trityl-protected amino-modifiers.2 In DNA oligonucleotides, a nonvolatile base, such as TRIS, is added prior to drying down to prevent detritylation. However, the presence of TRIS inhibits the subsequent 2′-desilylation reaction with TEA•3HF for RNA.
The obvious next step was to test this on our RNA sequence:
5′-UUG UUC UUA UUG UUC UUA UU-3′
After synthesis on our ABI394 with TBDMS phosphoramidites, we used the following procedure:n
- Deprotect oligonucleotide as necessary according to nucleobase protecting groups and support.
- Add 45 mg Tris base/mL.
- Desalt on Glen-Pak™ DNA Cartridge.
- Condition cartridge with 0.5 mL ACN, followed by 1 mL 2M TEAA. \n
- Load oligonucleotide drop-wise onto Glen-Pak DNA Cartridge.
- Rinse with 2 mL RNase-free water.
- Rinse twice with 2 mL 0.5M aqueous sodium hydroxide.*
- Rinse with 2 mL RNAse-free water.
- Elute with 1 mL 75% ACN/RNase-free water.
- Dry down.
- 2′-Desilylation.
- Dissolve in DMSO (115 μL) and warm in a 65 °C water bath until fully dissolved.
- Add TEA (60 μL) and TEA•3HF (75 μL).
- Heat in a 65 °C water bath for 2.5 h.
- Quench with RNA quenching buffer (750 μL).
- Desalt on Glen Gel-Pak™ following recommended conditions and elute crude oligonucleotide in 0.1M RNase-free TEAA
The resulting oligonucleotide was analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC).
*In general, NaOH treatment is rather harsh for RNA oligonucleotides and we don’t typically see recommendations like this. Due to the short contact time while on the column, the sodium hydroxide did not negatively impact the RNA integrity, confirmed by the chromatograms. However, it is worth pointing out that this step should be done quickly. While we did not test other possibilities, alternative sodium-based solutions may work just as well to convert the oligonucleotide to the sodium salt on the Glen-Pak cartridge (e.g. 0.1M NaOH, 0.5M NaCO₃, or 0.5M NaHCO₃).
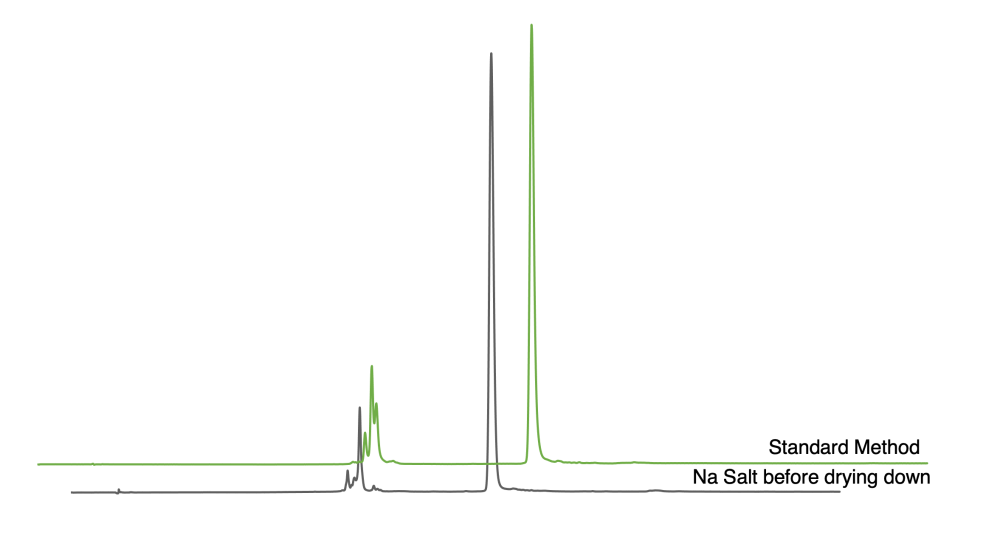
Figure 1. HPLC of 20 mer RNA oligonucleotides from TBDMS monomers with standard method (green) or converting oligonucleotide to the sodium salt before drying down (gray).
Following this method, we observed much better results by HPLC (Figure 1). In our standard method (green), the RNA was dried down prior to 2′-desilylation and desalting, and we saw failure sequences and a DMT-off full-length peak that eluted with the failures, according to MS analysis. However, when the RNA was converted to the nonvolatile sodium salt (gray) prior to drying down, we still saw the failure sequences but the DMT-off full length was significantly reduced. The effective average coupling efficiency was improved by 0.5%.
An alternative comparison method is to look at the area under the main peak. Converting the oligonucleotide to the sodium salt increased the percent area of the main peak by almost 10%. Explicitly, the main peak in the standard method totaled 77% and the main peak in the method evaluated in this article was 85%.
While we did not test this on TOM oligonucleotides, we previously observed the same loss of DMT using TOM phosphoramidites and expect this method to equally improve the overall coupling efficiency and yield of the DMT-ON full-length species.
References
- The Glen Report, 2024, 36.1, 9-13.
- The Glen Report, 2009, 21.1, 16.
- Glen Report 36-21: Spirocyclopropylene bridged nucleic acaid (scpBNA™) Phosphoramidites
- Glen Report 36-22: New Products Spirocyclopropylene Bridged Nucleic Acids
- Glen Report 36-23: Application Note Polymerase Chain Reaction (PCR) Optimization
- Glen Report 36-24: New Products Catalyst-free Click Reactions
- Glen Report 36-25: New Product Palmitate Serinol Phosphoramidite
- Glen Report 36-26: Technical Note Preventing Detritylation During RNA Deprotection
- Glen Report 36-27: Technical Snippets

