Glen Report 30.24: Oligonucleotide Dendrimers – An Update
Introduction
In a previous Glen Report article, Misha Shchepinov reviewed the variety of strategies available for the preparation and use of oligonucleotide dendrimers.1 In that article, he defined dendrimers as discrete, highly branched, monodispersed polymers that possess patterns reminiscent of the branching of trees. He also postulated that dendrimer oligonucleotides would be representative of a new segment of polymer science.
|
|
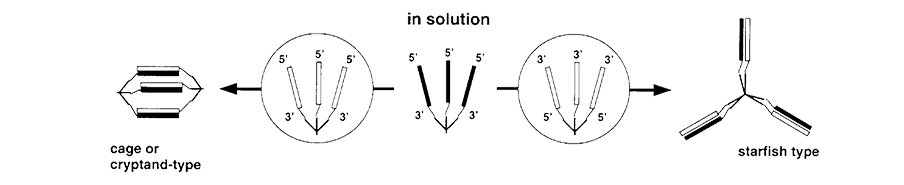
Figure 1. Formation of Complementary Dendritic Complexes |
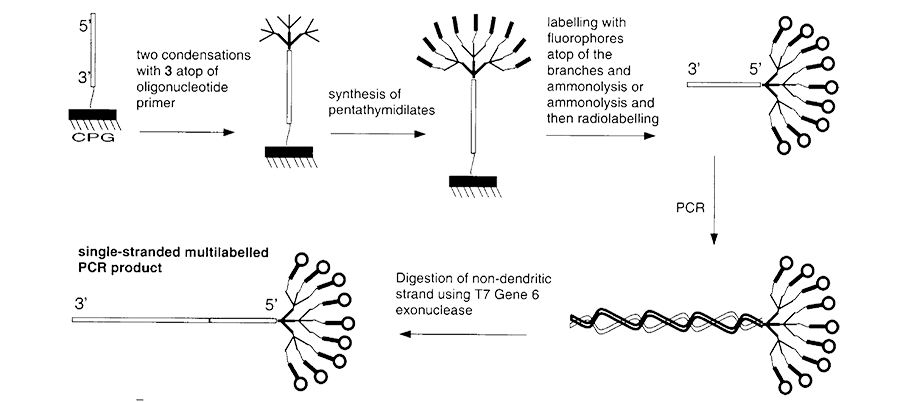
DNA molecules are well suited for use in nanotechnology because of their unique molecular recognition features. For example, dendrimers with arms terminating in oligonucleotides of the same or of different sequences could be used to build cages, cryptands, tubes, nets, scaffolds and other more complex 3-D structures (Figure 1).2,3 In the field of oligonucleotide arrays, where the signal is originally limited by the surface density of the device, the multiplicity of labeling afforded by dendritic structures allows detection at much lower concentrations. In the case of fluorescence detection, these structures allow amplified detection with adequate separation of labels available to avoid self-quenching. Similarly, dendritic labeling of probes allows for amplification of signal without affecting the behavior of the core oligonucleotide probe sequence (Figure 2).
|
|
Figure 2. Conversion of Polylabeled PCR Primer into a Single-Stranded Polylabeled PCR Product |
Glen Research offers a set of phosphoramidites for the synthesis of oligonucleotide dendrimers. These phosphoramidites, shown in Figure 4, Page 11, allow the dendritic segment to be readily prepared without affecting the structure of the core oligonucleotide.
Symmetrically Branched Dendrimers
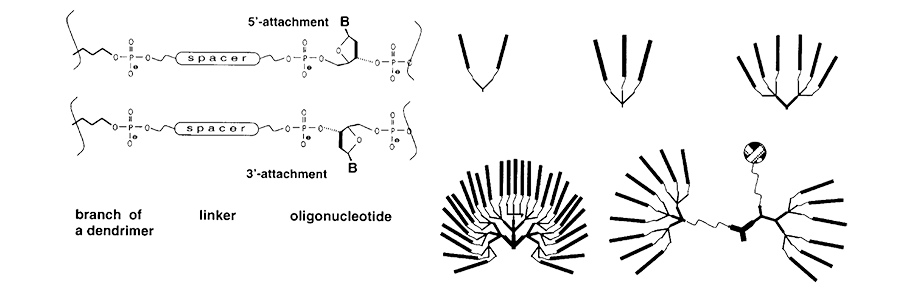
Symmetrically branched structures can be readily prepared using Symmetric Doubler (1), which simply doubles the branches in the growing oligonucleotide chain with every addition, as the name implies. More complex branches can be prepared by trebling the 5’ termini using Trebler (2) and Long Trebler (3), as illustrated in Figure 3.
|
|
Figure 3. Examples of Plain and Mixed Dendrimers of Different Generations |
As you can imagine, coupling branching species to the end of a growing oligonucleotide will potentially lead to the synthesis grinding to a halt for steric reasons. This was one of the reasons for adding Long Trebler (3) to the set since this molecule incorporates longer spacer arms in an attempt to alleviate steric effects. Regardless, the addition of intermediate spacer molecules, such as Spacer 18, is highly recommended.
As the density of oligonucleotides increases on the solid support, steric hindrance causes reactions to slow so complex dendritic oligonucleotides may need some optimization of the synthesis cycles. Certainly, the synthesis should be carried out on at least 1000Å CPG and preferably 2000Å CPG for more complex dendrimers.
Asymmetrically Branched Dendrimers
By far the most common approach to DNA diagnostics is amplification of the target sequence to produce enough copies for a signal to be observed using conventional detection systems. However, an alternative approach does exist - direct analysis of the target DNA by signal amplification.4-7 This latter technique requires that the synthetic oligonucleotide should contain the primary sequence attached to many identical copies of the secondary sequence. It is the detection of the many copies of the secondary sequence that serves to amplify the signal. The resulting branched oligonucleotide has been aptly described as comb-like, with the primary sequence in the handle and the secondary sequences being the teeth of the comb. The synthesis of comb-like structures requires the use of an asymmetric branching phosphoramidite.
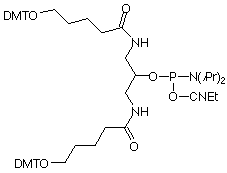 |
|
Symmetric Doubler (1) |
Trebler (2) |
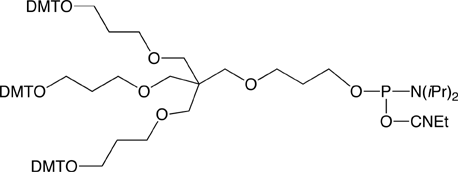 |
|
Long Trebler (3) |
|
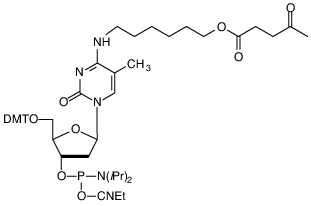 |
 |
5-Me-dC Brancher (4) |
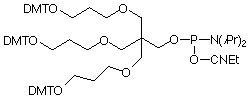
Asymmetric Doubler (LEV) (5) |
Figure 4. Structures of Symmetric and Assymetric Branchers |
|
Glen Research previously offered an Asymmetric Brancher with the site destined to become part of the comb sequence blocked with the ubiquitous Fmoc protecting group.8 Unfortunately, this structure proved to have limited stability as a phosphoramidite and was discontinued a few years ago. Following the success and inherent stability of the 5-Me-dC Brancher Phosphoramidite (4), we have now introduced Asymmetric Doubler Lev (5), also protected with a levulinyl (Lev) protecting group. Once the primary oligonucleotide sequence ‘handle’ of the comb structure has been synthesized, the Lev protecting group can be specifically removed with a hydrazine reagent and the oligonucleotide ‘teeth’ can then be synthesized.
Synthesis of these comb structures has been described in detail for dC Brancher and is included on Page 2 of its Technical Bulletin - Synthesis of branched DNA with a comb structure.
References
1. The Glen Report, 1999, 12, 1-4.
2. M.S. Shchepinov, I.A. Udalova, A.J. Bridgman, and E.M. Southern, Nucleic Acids Res, 1997, 25, 4447-4454.
3. M.S. Shchepinov, K.U. Mir, J.K. Elder, M.D. Frank-Kamenetskii, and E.M. Southern, Nucleic Acids Res, 1999, 27, 3035-41.
4. T. Horn, and M.S. Urdea, Nucleic Acids Res, 1989, 17, 6959-67.
5. M.L. Collins, et al., Nucleic Acids Res, 1997, 25, 2979-84.
6. T. Horn, C.A. Chang, and M.S. Urdea, Nucleic Acids Res, 1997, 25, 4835-4841.
7. T. Horn, C.A. Chang, and M.S. Urdea, Nucleic Acids Res, 1997, 25, 4842-4849.
8. M.S. Shchepinov, and E.M. Southern, Russ. J. Bioorg. Chem., 1998, 24, 794.