Glen Report 21.21: 5-Hydroxymethyl-dC: A New Actor in the Field of Epigenetics
One of the fastest growing fields in biology and cancer research is epigenetics. While the underlying genetic code defines which proteins and gene products are synthesized, it is epigenetic control that defines when and where they are expressed. This dynamic control of gene expression is essential for X chromosome inactivation, embryogenesis, cellular differentiation and appears integral to memory formation and synaptic plasticity.1,2
Epigenetic control is generally mediated by methylation of cytidine to 5-methyl-dC in CpG sites and post-translational modification of histones. Methylation of CpG sites near promoters is associated with gene silencing, as is deacetylation of histones. A number of diseases can result when CpG methylation control is lost or when the de novo methylation is incorrect, such as Rett, Fragile X, ATR-X, Prader-Willi and Angelman syndromes. In addition, dysregulated methylation patterns are often seen in cancers and this is widely thought to contribute to tumorigenesis.3
While a number of mammalian DNA methyltransferases are known in the DNMT family, the enzymes responsible for demethylation in mammals have yet to be identified conclusively since none has shown clear activity in vitro.4 However, recent work by Skirmantas Kriaucionis in the Heintz Lab. demonstrated that 40% of the purported 5-methyl-dC in Purkinje neurons was actually 5-hydroxymethyl-dC.5 This hydroxymethyl cytidine analog has low affinity for the Methyl-CpG-binding Protein MeCP2, which is a known transcriptional repressor, as well as DNMT1, which is the maintenance DNA methyltransferase.6,7 This opens up the possibility that demethylation would be acquired passively over multiple cell cycles if a means could be found to convert 5-methyl-dC to 5-hydroxymethyl-dC. Less than a month after Kriaucionis’ publication, the Rao Lab. found an enzyme, TET1, which catalyzes the conversion of 5-methyl-dC to 5-hydroxymethyl-dC in vitro and in vivo, ushering in a new chapter in the field of epigenetics with 5-hydroxymethyl-dC taking center stage.8
Another consideration is the potential for 5-hydroxymethyl-dC to be a bad actor in DNA damage. After all, 5-hydroxymethyl-dU is a product of oxidative damage of thymidine by hydroxyl radicals or ionizing radiation. Similarly, 5-hydroxymethyl-dC may be a damaged form of 5-methyl-dC. However, it is unlikely that the 5-hydroxymethyl-dC detected in neuronal cells is formed by oxidative damage since no other damaged nucleosides, e.g., 8-oxo-dG, were detected in these cells.5 Nevertheless, 5-hydroxymethyl-dC has been detected in bacteria9 and it is certainly feasible that it could still be a product of oxidative damage in mammalian cells.
With a growing audience for all aspects of 5-hydroxymethyl-dC activity, it seems useful to offer this nucleoside as a phosphoramidite. The protection scheme described by Sowers9 has performed well for oligonucleotide synthesis in our hands. With benzoyl protection of the 5-hydroxymethyl-dC residue, this monomer is not compatible with methylamine deprotection or UltraMild chemistry. However, deprotection is clean and complete with ammonium hydroxide at 30% Ammonium Hydroxide for 17 hours at 75°C.
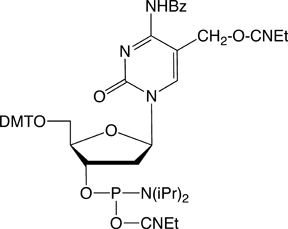
5-Hydroxymethyl-dC
References
- J. Turek-Plewa, and P.P. Jagodzinski, Cell Mol Biol Lett, 2005, 10, 631-47.
- C.A. Miller, S.L. Campbell, and J.D. Sweatt, Neurobiol Learn Mem, 2008, 89, 599-603.
- K.D. Robertson, Nat Rev Genet, 2005, 6, 597-610.
- S.K. Ooi, and T.H. Bestor, Cell, 2008, 133, 1145-8.
- S. Kriaucionis, and N. Heintz, Science, 2009, 324, 929-30.
- V. Valinluck, et al., Nucleic Acids Res., 2004, 32, 4100-8.
- V. Valinluck, and L.C. Sowers, Cancer Res, 2007, 67, 946-50.
- M. Tahiliani, et al., Science, 2009, 324, 930-5.
- S. TardyPlanechaud, J. Fujimoto, S.S. Lin, and L.C. Sowers, Nucleic Acids Res., 1997, 25, 553-558.
Product Information
- Glen Report 21.21: 5-Hydroxymethyl-dC: A New Actor in the Field of Epigenetics
- Glen Report 21.22: Purification of CleanAmp™ DNA Oligonucleotides (DMT-ON)
- Glen Report 21.23: A 3'-Cap for Improved Target Affinity and Specificity
- Glen Report 21.24: Metallobase Nucleic Acid Modification
- Glen Report 21.25: tC and tCo: New Tricyclic Fluorescent Cytidine Analogues with a very Bright Future
- Glen Report 21.26: Non-Aqueous Oxidation for PACE Chemistry
- Glen Report 21.27: New Product - Azobenzene Phosphoramidite for the Introduction of Photo-regulated Functions in DNA
- Glen Report 21.28: Deprotection - Volume 3 - Dye-Containing Oligonucleotides
- Glen Report 21.29: New Products - Glen UnySupport™ Frits
- Glen Report 21.210: New products: Glen-Pak™ DNA Cartridge 3g and DNA 30mg 96-Well Plates
- Glen Report 21.211: Technical Brief – Synthesis of Long Oligonucleotides

