Glen Report 20.110: Improving Universal Support II for Oligonucleotide Synthesis
In 2001, Azhayev and his group at the University of Kuopio designed and reported1 on several new universal supports for oligonucleotide synthesis, which lack the limitations of similar and previously reported solid phases.2-4 Several properties of these new solid phases make them rather attractive in industrial high output multi-well synthesizers. Our results have demonstrated that these new matrices are:
- Fast: cleavage and dephosphorylation in 20 minutes at room temperature;
- Mild: cleavage reagent is 2 M ammonia in methanol;
- Compatible with UltraMild, normal, and UltraFast deprotection; and
- Cost-effective: comparable in price to regular 2’-deoxynucleoside supports.
Scheme 1 describes a procedure for the preparation of USII support. Initially, long chain alkylamino controlled pore glass (lcaaCPG) or Macroporous Aminomethyl-polystyrene is succinylated and capped to give succinylamido-support (1). (±)-3-Amino-1-(4,4'-dimethoxytriphenylmethyl)-2-propanediol (2) is obtained in 3 steps and then attached to support (1) in the presence of N,N’-di-isopropylcarbodiimide and N-hydroxybenzotriazole to give solid support (3). The unreacted carboxyl groups of support (3) are capped with benzylamine, and only then the free hydroxyl group of support (4) is dichloroacetylated, to give the final support, Universal Support II (US II).
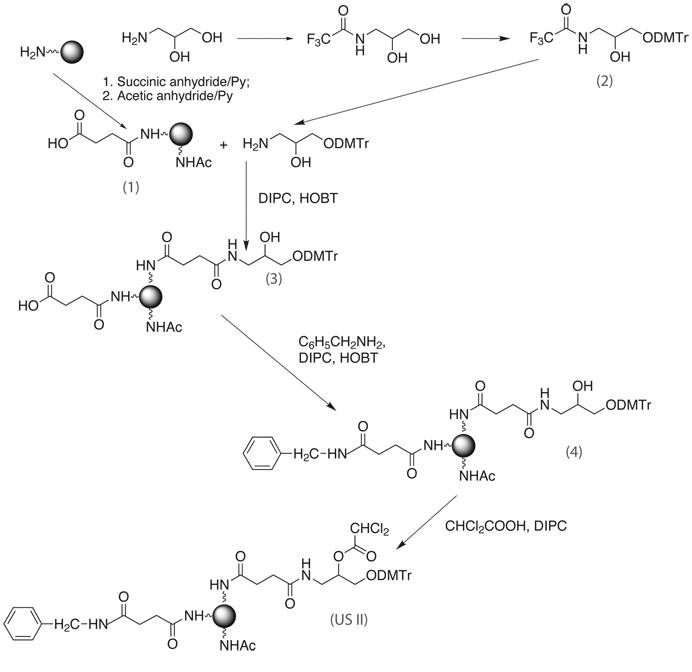
R=Cl2CH-; DIPC: N,N'-di-isopropylcarbodiimide; HOBT: N-hydroxybenzotriazole; Support = aminomethyl-polystyrene or long chain alkylamino-CPG.
The preparation of the linker precursor (2) consists of 3 steps. The synthesis of US II, therefore, consists of 6 steps, 3 of which take place on solid phase.
Scheme 2 shows a procedure for the preparation of the new Universal Support III (US III). The Carbomoylation Linker (5)for US III preparation is synthesized in 3 simple steps in 80-85% yield and then directly attached to the Macroporous Aminomethyl-polystyrene, employing a new carbomoylation procedure.5 Finally, the unreacted aminomethyl groups of the Aminomethyl-polystyrene are capped with dichloroacetyl imidazole to give the USIII support.
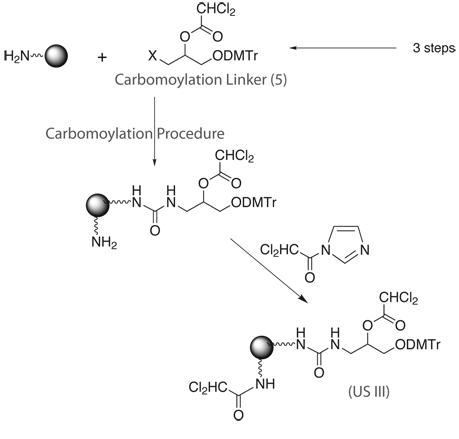
Support = aminomethyl-polystyrene.
Therefore, the preparation of the Carbomoylation Linker consists of 3 steps. And the preparation of US III takes only 2 steps of modification on solid phase – attachment of linker and a capping step.
The synthesis of US II is a lengthy and laborious procedure - 3 steps to the linker (2) and 3 further reactions on solid phase. Preparation of US III, comprising only 2 steps of solid phase modification, is a much more straightforward procedure. Especially noteworthy is the fact that the required loading of the Carbomoylation Linker, (5) in Scheme 2, on solid phase is much easier to achieve when using the carbomoylation procedure.
Taken together, these facts make US III an improved product when compared to US II. Moreover, the new US III appears the same as, if not better than, US II in terms of performance as the truly universal solid support for oligonucleotide synthesis.
Because the universal linker is unchanged and the succinate or urea groups remain attached to the support, we use the same catalog numbers for US II and III. Using Universal Support II or III, an oligo yield of > 80% can be achieved on CPG supports and > 95% on polymeric supports, with purity equivalent to the same oligo prepared normally.
In addition, the carbomoylation process using the Carbomoylation Linker, (5) in Scheme 2, is now available for license to those who would prefer to produce US III on their own solid matrices.
References
- A.V. Azhayev, and M.L. Antopolsky, Tetrahedron, 2001, 57, 4977-4986.
- S. Scott, P. Hardy, R.C. Sheppard, and M.J. McLean A universal support for oligonucleotide synthesis, In Innovations and Perspectives in Solid Phase Synthesis, 3rd International Symposium, 1994; R. Epton, Ed. Mayflower Worldwide: 1994; pp 115-124.
- M.H. Lyttle, D. Hudson, and R.M. Cook, Nucleic Acids Res, 1996, 24, 2793-8.
- A.V. Azhayev, Tetrahedron, 1999, 55, 787-800.
- The new carbomoylation chemistry, resulting in the stable urea fragment bridging the universal linker and aminoalkylated solid phase, is subject to proprietary rights of Glen Research Corporation and Metkinen Chemistry (U.S. Patent Application Serial No 60/854,721; International Patent Application No. PCT/FI2007/050575). Universal Support II and III are covered by the following intellectual property: US Patent No.: 6,770,754 and European Patent No.: 1404695.
What are the structural differences between US II and US III?
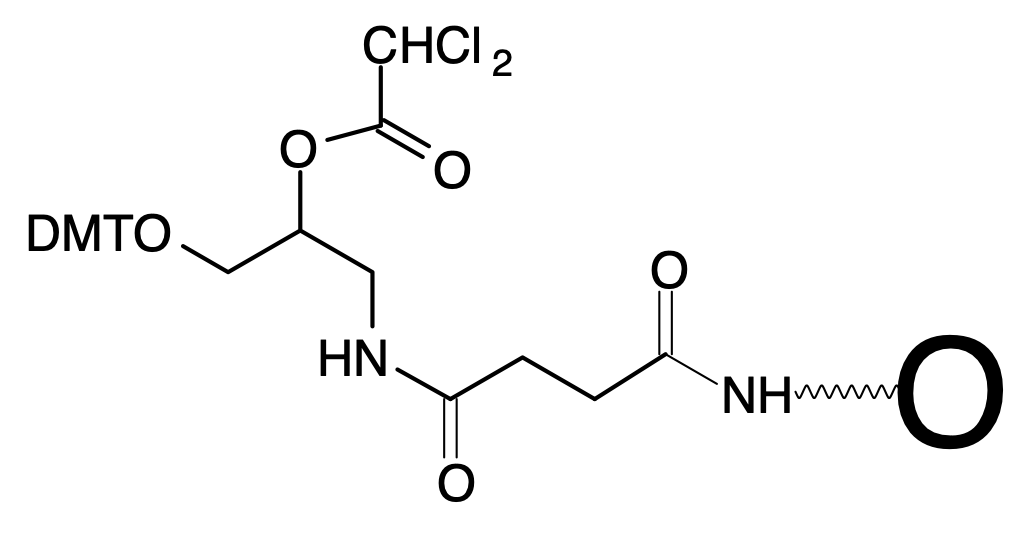
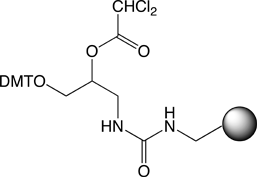
Differences Between Universal Supports II and III
The universal linker, the top left section of structures 1 and 2 in Figure 1, is identical in Universal Support II and Universal Support III. The difference is in the attachment of the universal linker to the support. In the case of Universal Support II, the attachment is through a succinoyl diamide linkage while in Universal Support III it is a urea linkage. The amide assisted dephosphorylation reaction that releases the oligo-3'-OH into solution takes place in the universal linker section and the remainder of the attachment remains with the support after release of the oligonucleotide. So the structural differences have no effect on the function of the two supports and have no impact on the product oligonucleotide.
Why was the change necessary?
The synthetic changes described in this article were made to improve the control of the production of the universal support. In the case of Universal Support II, three reactions on solid phase are required and the quality analysis can only be carried out after these three steps. In the case of Universal Support III, the loading reaction is followed by simple capping of the support. The support can be analyzed after both steps. Clearly, the production of Universal Support III is under much more control and is also more amenable to scale up.
Why are the catalog numbers the same?
We have retained the same catalog numbers for Universal II and III since the difference only affects the support. (The situation is similar to lcaa-CPG, the core of most CPG supports, where individual manufacturers may use different structures for the long chain alkylamine (lcaa) linker.) All of our polymeric supports of this type have already been changed to the Universal Support III structure and the name and structure on the analytical data reflect this change. In future, CPG supports may also be changed if the benefits of the change are the same for CPG as for polymeric supports.
Can I make my own support?
The Carbomoylation Linker, (5) in Scheme 2, will be available commercially very shortly. The carbomoylation chemistry used to make the urea linkage is proprietary and details will be released as soon as the patent process is complete. In the meantime, the technology, Universal Support III and the carbomoylation technology required to produce the supports is available for licensing. Contact support@glenres.com for more information.
Has the performance of the support been affected?
Universal Support III performs identically to Universal Support II. However, since tighter control of production is possible, Universal Support III will always perform at the highest level previously achieved by Universal Support II.
Has the change affected my cost?
Cost is always paramount in everyone's mind when related to high throughput or large-scale oligonucleotide synthesis. The much improved synthesis of Universal Support III using the carbomoylation procedure also improved the cost structure. Universal Support III can be offered at about the same price as regular 2'-deoxynucleoside supports and at a lower price than ribonucleoside supports. Please contact us for a quotation.
Product Information
Universal Support II has been discontinued. Please see:
- Glen Report 20.11: An Unnatural Base Pair System for the Expansion of Genetic Information
- Glen Report 20.12: Thiophosphoramidites and Their Use in Synthesizing Oligonucleotide Phosphorodithioate Linkages
- Glen Report 20.13: Technical Brief - Purification of 6-FAM Labelled oligos using Glen-Pak™ Cartridges
- Glen Report 20.14: More Click Chemistry
- Glen Report 20.15: Technical Brief - New application of 5-Me-iso-dC and iso-dG
- Glen Report 20.16: Technical Brief - New application for 5’-OMe-dT phosphoramidite
- Glen Report 20.17: New Product - Deuterated 2’-Deoxyguanosine Phosphoramidite
- Glen Report 20.18: New Products - Solid CPR II / Formylindole – Aldehyde Modifier
- Glen Report 20.19:New Product - 5'-Cholesteryl-TEG Phosphoramidite
- Glen Report 20.110: Improving Universal Support II for Oligonucleotide Synthesis
- Glen Report 20.111: Differences Between Universal Support II and III
- Glen Report 20.112: Technical Brief - Phage Display, Artificial Antibodies and Trimer Phosphoramidites

