Glen Report 26.21: Reversible M6A RNA Modification
Qing Dai and Chuan He
Department of Chemistry and Institute for Biophysical Dynamics
The University of Chicago
929 East 57th Street
Chicago, Illinois 60637, USA
In the central dogma of molecular biology, genetic information flows from DNA to RNA and then to protein. Reversible epigenetic modifications on genomic DNA1 and histone2 have been known to substantially regulate gene expression (Figure 1). On the other hand, there exists more than 100 naturally occurring chemical modifications in RNA3; however, the functions of these RNA modifications are largely unknown. Whether some of these modifications in RNA can be reversed and could impact gene expression in the central dogma was unknown until the recent discovery of N6-methyladenosine (m6A) as the first example of reversible RNA methylation4.
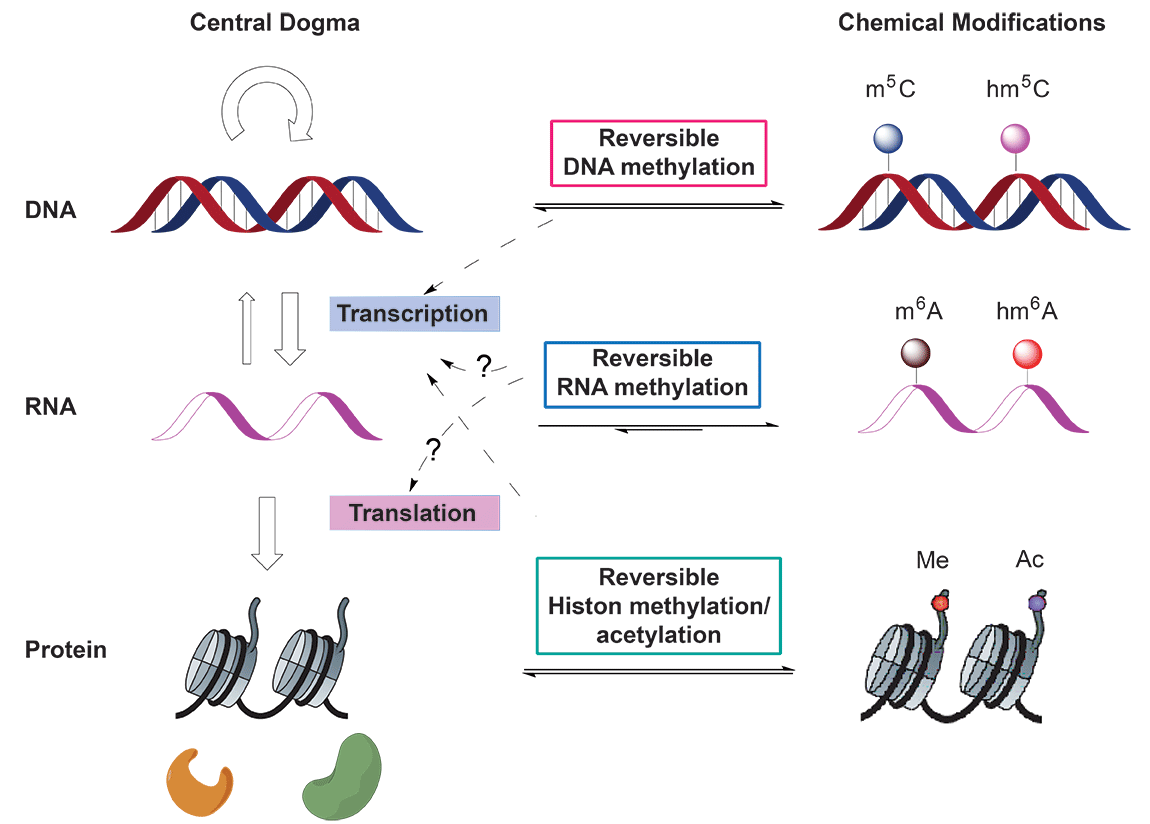
(Adapted from Y. Fu et al., Nat. Rev. Genet. 2014, 15, 293-306)
Discovered in the 1970s5, the m6A modification exists in different types of RNA including rRNA, snRNA, tRNA, mRNA, and lncRNAs etc. It is the most prevalent internal modification in mRNAs and lncRNAs in higher eukaryotes6. The amount of m6A in isolated RNA was estimated to be 0.1–0.4% of that of adenines (that is, ~3–5 m6A sites per mRNA) in mammals and the identified consensus sequence is [G/A/U][G>A]m6AC[U>A>C]7. The genome-wide distribution of m6A in mammals was recently revealed using m6A-seq by two laboratories8. The resulting maps have shown that m6A is widely distributed in more than 7,000 mRNA and 300 non-coding RNA (ncRNA) transcripts in human cells and mouse brain. m6A is also enriched around stop codons, in 3' untranslated regions (3'UTRs), and within internal long exons. Many m6A peaks are well conserved between humans and mice, and dynamic changes of certain peaks have been observed under different stress conditions. These observations further suggest that the m6A modification in mRNA and lncRNA may have significant biological functions.
In 2011, the discovery of the α-ketoglutarate-dependent dioxygenase FTO as the first RNA demethylase reignited investigations of m6A biology4a. FTO is an intriguing protein that has been associated with human obesity. This protein efficiently demethylates m6A via two unprecedented intermediates, N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A), which were generated through the FTO-catalyzed oxidation of m6A (Figure 2)9. Further experiments showed that silencing of FTO in HeLa and 293FT cells increased the total m6A level in polyadenylated RNA, where as over-expression of FTO decreased the total m6A level in polyadenylated RNA. Subsequently, ALKBH5 was found to be the second demethylase that shows efficient demethylation activity towards m6A in mRNA and other nuclear RNA4b. The reversible nature of the m6A modification and the unique distribution pattern of m6A in mRNA have attracted great attention from the biological community since.
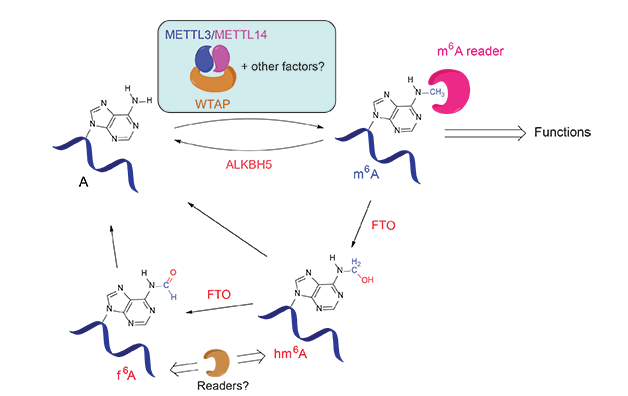
(Adapted from Y. Fu et al., Nat. Rev. Genet. 2014, 15, 293-306)
It has been known that METTL3 is an active component of the m6A methyltransferase complex in mammalian cells but other components are required to achieve optimal activity in vitro. In 2014, METTL14 was characterized to be another active component of the m6A methyltransferase complex and forms a stable hetero complex with METTL310 (Figure 2). Biochemical characterization revealed that these two proteins form a stable complex with a stoichiometric ratio of 1:1. The combination of both methyl transferases leads to a substantially enhanced methylation activity. This heterodimer also shows a strong preference for the cognate m6A consensus sequence and a modest preference for less structured RNA in vitro. WTAP, a protein involved in splicing, is the third critical component of the m6A methyltransferase complex in vivo10a,11 (Figure 2).
For the m6A group to have a biological function, it needs to be recognized through “reading” by specific proteins. This process could resemble the roles of proteins that read 5-methylcytosine (5mC) in DNA, or methylated or acetylated amino acid residues of histones in order to exhibit biological functions associated with the modifications and to enable reversible tuning. Pull-down experiments using synthetic RNA probes in the presence and absence of m6A identified the YTH family proteins as m6A-specific binding proteins8a,12. One of the reader proteins, YTHDF2, preferentially recognizes m6A-containing mRNA and regulates the degradation of the methylated transcripts12 (Figure 2). A recent structural characterization revealed a well-conserved hydrophobic pocket used by the YTH family proteins to selectively recognize the methyl group of m6A in RNA13.
The discoveries and characterization of the m6A writers, erasers and readers, together with the parallel development of high-throughput assays that profile this methylation on a transcriptome-wide scale, set the stage and provided tools for functional investigations that aim to identify the mechanisms by which m6A is translated into biological outcomes.
The synthesis of m6A phosphoramidite and its incorporation into RNA have helped to discover the reader and eraser proteins and will find more applications in studies on how m6A affects RNA structures, recruits binding proteins, and interacts with writers and erasers. It may also find applications on developing inhibitors to interfere with the m6A writing, erasing, and reading processes as potential drug candidates.
The synthesis of the first version of m6A phosphoramidite (I) without protecting the MeNH- group has been reported14 (Figure 3). This version of m6A phosphoramidite couples as efficiently as unmodified phosphoramidites when tetrazole is used as the activator. After deprotection, HPLC analysis showed the full-length RNA to be the major product. It is possible that the unprotected MeNH- group may also be subject to coupling to give a branched RNA byproduct, especially when DCI is used as the activator.
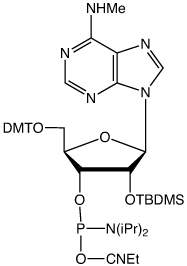
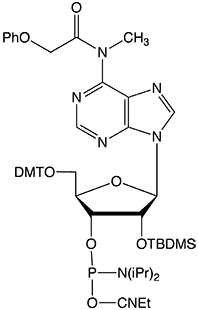
To completely eliminate the possible coupling of MeNH- to form an undesired byproduct, Glen Research has developed a new version of m6A phosphoramidite with MeNH- protected by the PhOAc group (II). This protecting group can be readily removed under normal base treatment during deprotection. The availability of this new m6A phosphoramidite will significantly facilitate investigations of m6A and its related biological functions.
References
-
- M. M. Suzuki, et al., Nat. Rev. Genet., 2008, 9, 465–476.
- R. M. Kohli, et al., Nature, 2013, 502, 472–479.
- P. A. Jones, Nat. Rev. Genet., 2012, 13, 484–492.
- M. R., Branco, et al., Nat. Rev. Genet. 2012, 13, 7–13.
- N. Bhutani, et al., Cell, 2011, 146, 866–872.
-
- B. D. Strahl, et al., Nature, 2000, 403, 41–45.
- Y. Shi, Nat. Rev. Genet., 2007, 8, 829–833.
- R. J. Klose, et al., Nat. Rev. Genet., 2006, 7, 715–727.
- A. Bird, Science. 2001, 294, 2113–2115.
- M. A. Machnicka, et al., Nucleic Acids Res. 2013, 41, D262–D267.
-
- G. Jia, et al., Nat. Chem. Biol. 2011, 7, 885–887.
- G. Zheng, et al., Mol. Cell, 2013, 49, 18–29.
-
- R. P. Perry, et al., Cell, 1974, 1, 37–42.
- R. Desrosiers, et al., Proc. Natl. Acad. Sci. USA, 1974, 71, 3971–3975.
- C. M. Wei, et al., Cell, 1975, 4, 379–386.
-
- P. Narayan, et al., Science, 1988, 242, 1159–1162.
- T. Csepany, J. Biol. Chem., 1990, 265, 20117–20122.
- P. Narayan, et al., Nucleic Acids Res., 1994, 22, 419–426.
-
- D. Dominissini, et al., Nature, 2012, 485, 201-208.
- K. D. Meyer, et al., Cell, 2012, 149, 1635–1646.
- Y. Fu et al., Nature Commun., 2013, 4, 1798.
-
- J. Liu, et al., Nature Chem. Biol., 2014, 10, 93-95.
- Y. Wang, et al., Nat. Cell Biol., 2014, 16, 191–198.
- X. L. Ping, et al., Cell Res. 2014, 24, 177–89.
- X. Wang et al., Nature, 2014, 505, 117–20.
- C. Xu, et al., Nat. Chem. Biol. 2014, 10, 927–9.
- Q. Dai et al., Nucleic Acids Res., 2007, 35, 6322–6329.
Product Information
- Glen Report 26.21: Reversible M6A RNA Modification
- Glen Report 26.22: New Product - N6-Me-A -Reversible M6A RNA Modification
- Glen Report 26.23: New Product - 5'-Carboxy-Modifier C5
- Glen Report 26.24: DNA-Mediated Charge Transport: A Novel Property of DNA with Diverse Applications
- Glen Report 26.25: New Product - Methylene Blue NHS Ester
- Glen Report 26.26: Technical Brief - Which 3'-Amino-Modifier?
- Glen Report 26.27: Technical Brief – Use of Click Chemistry for Possible Crosslinking, Click Synthesis of a Potentially Useful azido support and New Click Product - Cyanine 7 Azide
- Glen Report 26.28: New Product - Disulfo-Cyanine 7 Azide

