Glen Report 30.11: Introducing ribo-tCo<>/sup>
Authors: Anders Foller Füchtbauer and Marcus Wilhelmsson
Department of Chemical and Biological Engineering/Physical Chemistry
Chalmers University of Technology
Kemivägen 10, SE-412 96 Göteborg, Sweden
Figure 1. Guanine base paired with tC
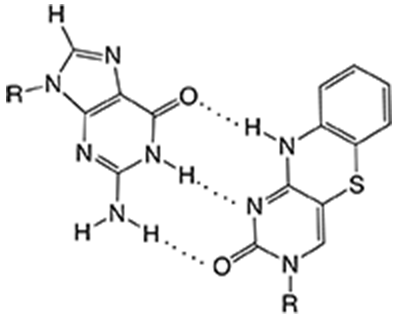
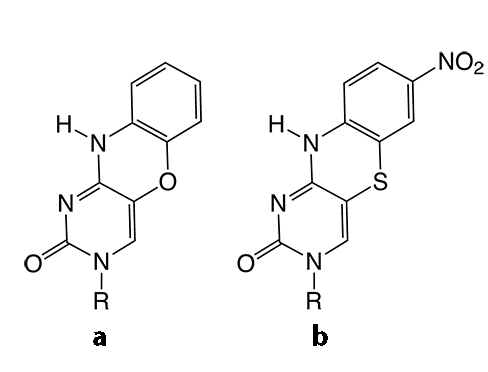
The bright fluorescent tricyclic cytosine analogues tC (Figure 1) and tCO that we have developed and that Glen Research has successfully supplied since 2009, stand out among fluorescent bases due to their virtually unquenched fluorescence inside single- or double-stranded DNA.1-3 Until recently the family of tricyclic cytosines had only been studied1-3 and used4-12 in DNA contexts and, importantly, introduced as possible donors of the first DNA base analogue FRET-pair with tCnitro (Figure 2, Page 2).4
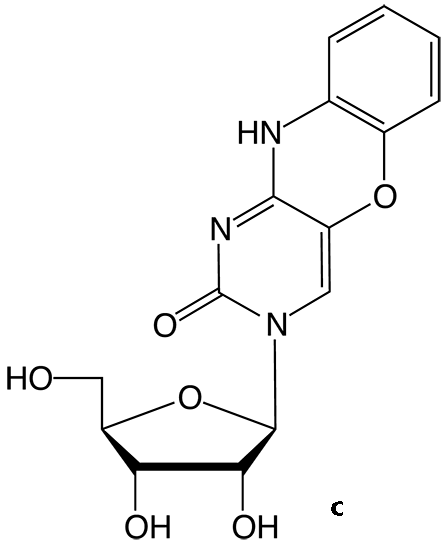
However, in 2017, we also reported on the synthesis of the tCO ribonucleoside (Figure 2c) and its incorporation into a range of RNA sequences, where it was shown to be a very potent and useful fluorophore also in this context.13 Therefore, Glen Research in cooperation with ModyBase HB has decided to offer this useful fluorescent ribonucleoside analogue.
Figure 2. Structures of tricyclic cytosine analogues tCO (a) tCnitro (b) and the tCO ribonucleoside (c).
 |
 |
Fluorescent base analogues for RNA are limited in number compared to their DNA counterparts. To facilitate the application of such analogues, characterization of their structural and dynamics behavior in RNA compared to the corresponding natural nucleoside is important. Moreover, characterization of their photophysical properties as an effect on various surrounding RNA bases is vital. However, such thorough characterizations inside RNA are rare. Therefore, when we reported on the first synthesis and RNA-incorporation of the tCO-ribonucleoside and characterized its base-mimicking and fluorescence properties in RNA in 2017 this was one of the first such thorough investigations.13 The tCO-ribonucleoside, like its deoxy-counterpart, can be selectively excited with a peak around 365 nm well outside the nucleobase region and has a broad emission band with a peak, slightly dependent on sequence context, at approximately 455 nm (see Figure 3, Page 2 and Table 1, Page 2).
Measurements were performed in PBS buffer (100 mM Na+, pH 7.5).a. Sequences are named after the bases flanking tCO in the sequence 5'- CACX2X1tCOY1Y2CC - 3'.
b. Fluorescence quantum yields are measured relative to the quantum yield of the potassium salt of the tCO-monomer in water (φF = 0.30).3
c. Amplitude-weighted mean fluorescence lifetime, <τ> = Saiti/Sa
Table 1. Photophysical properties of the tCO monomer ribonucleoside and tCO in various RNA sequence surroundings. |
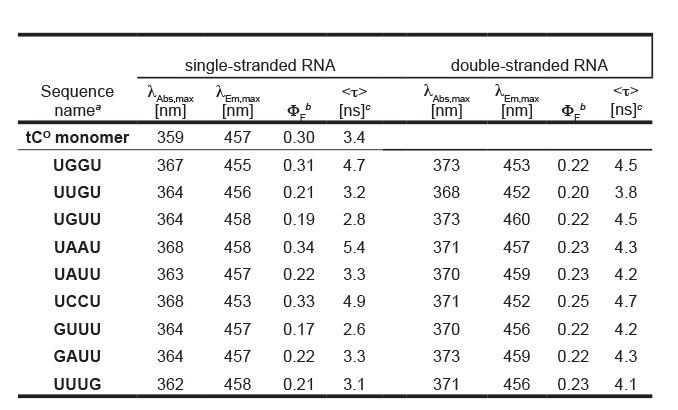 |
Figure 3. Normalized absorption and emission of tCO as a monomer ribonucleoside and inside single- and double-stranded RNA. |
 |
As in DNA, tCO displays high quantum yields inside RNA duplexes (<φF>=0.22) that are almost unaffected by neighboring RNA bases (Table 1). This results in an average brightness of 1900 M-1cm-1, which is significantly higher than previously reported fluorescent bases inside RNA.13
The average fluorescence lifetime of tCO inside RNA duplexes with different neighboring bases is 4.3 ns (see Table 1 for details) and in general two lifetimes are required to fit the exponential decays.13 The fluorescence properties in single-stranded RNA are characterized by a slight increase in average quantum yield (<φF>=0.24) compared to the corresponding duplex RNAs, with a broader distribution and somewhat shorter average lifetimes (Table 1).13 Importantly, using circular dichroism (CD) we also found that the tCO-modified RNA duplexes form regular A-helices and in UV-melting experiments the stability of the duplexes was found to be only slightly higher than that of the corresponding natural RNA (<ΔTm>=+2.3°C).13
Glen Research is therefore pleased to introduce the ribonucleoside of tCO (Ribo-tCO-CE Phosphoramidite, Figure 4) and believes that its properties make it a highly interesting and useful bright internal RNA label for a wide range of spectroscopy and microscopy experiments and also as an excellent analogue of cytosine in RNA.
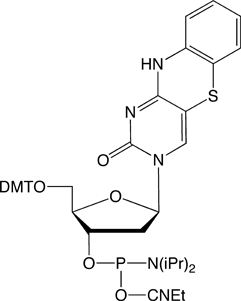
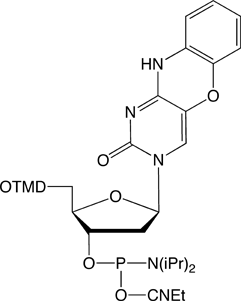
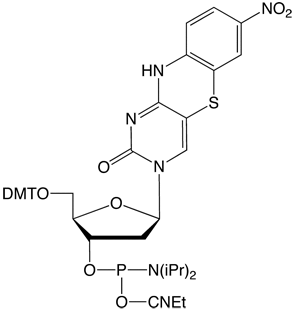
Figure 4. Structures of tricyclic cytosine analogue phosphoramidites. |
|||
 |
 |
 |
|
tC-CE Phosphoramidite (10-1516) |
tCO-CE Phosphoramidite (10-1517) |
tCnitro-CE Phosphoramidite (10-1518) |
Ribo-tCO-CE Phosphoramidite (10-3517) |
References
1. A Highly Fluorescent DNA Base Analogue that Forms floatLefttson-Crick Base Pairs with Guanine. Wilhelmsson L.M., Holmén, A., Lincoln, P., Nielsen, P.E., Nordén, B.. J. Am. Chem. Soc.; 2001, 123(10); 2434-2435.
2. Fluorescent Properties of DNA Base Analogue tC upon Incorporation into DNA – Negligible Influence of Neighboring Bases on Fluorescence Quantum Yield. Sandin, P., Wilhelmsson, L.M., Lincoln, P., Powers, V.E.C., Brown, T., Albinsson, B.. Nucleic Acids Res.; 2005, 33(16); 5019-5025.
3. Characterisation and Use of an Unprecedentedly Bright and Structurally Non-perturbing Fluorescent DNA Base Analogue. Sandin, P., Börjesson, K., Li, H., Mårtensson, J., Brown, T., Wilhelmsson, L.M., Albinsson, B.. Nucleic Acids Res.; 2008, 36(1); 157-167.
4. A Nucleic Acid Base Analog FRET-pair Facilitating Detailed Structural Measurements in Nucleic Acid Containing Systems. Börjesson, K., Preus, S., El-Sagheer, A.H., Brown, T., Albinsson, B., Wilhelmsson, L.M.. J. Am. Chem. Soc.; 2009, 131; 4288-4293.
5. Highly Efficient Incorporation of the Fluorescent Nucleotide Analogues tC and tCO by Klenow Fragment. Sandin, P., Stengel, G., Ljungdahl, T., Börjesson, K., Macao, B., Wilhelmsson, L.M.. Nucleic Acids Res.; 2009, 37(12); 3924-3933.
6. Ambivalent Incorporation of the Fluorescent Cytosine Analogues tC and tCO by Human DNA Polymerase α and Klenow Fragment. Stengel, G., Purse, B.W., Wilhelmsson, L.M., Urban, M., Kuchta, R.D.. Biochemistry; 2009, 48(31); 7547-7555.
7. Discrimination against the Cytosine Analog tC by Escherichia coli DNA Polymerase IV DinB. Walsh, J.M., Bouamaied, I., Brown, T., Wilhelmsson, L.M., Beuning, P.J.. J. Mol. Biol.; 2011, 409; 89-100.
8. Mammalian Transcription Factor A is a Core Component of the Mitochondrial Transcription Machinery. Shi, Y., Dierckx, A., Wanrooij, P.H., Larsson, N-G., Wilhelmsson, L.M., Falkenberg, M., Gustafsson, C.M..Proc. Natl. Acad. Sci. U.S.A.; 2012, 109(41), 16510-16515.
9. FRETmatrix: A General Methodology for the Simulation and Analysis of FRET in Nucleic Acids. Preus, S., Kilså, K., Miannay, F-A., Albinsson, B., Wilhelmsson, L.M.. Nucleic Acids Res.; 2013, 41(1), e18.
10. Studying Z-DNA and B- to Z-DNA transitions using a cytosine analogue FRET-pair. Dumat, B., Larsen, A.F., Wilhelmsson, L.M.. Nucleic Acids Res.; 2016, 44, e101.
11. Twist-open mechanism of DNA damage recognition by the Rad4/XPC nucleotide excision repair complex. Velmurugu, Y. Chen, X.J., Sevilla, P.S., Min, J.H., Ansari, A.. Proc. Natl. Acad. Sci. U.S.A.; 2016, 113, E2296-E2305.
12. In Vitro Fluorogenic Real-Time Assay of the Repair of Oxidative DNA Damage. Edwards S.K., Ono T., Wang S., Jiang W., Franzini R.M., Jung J.W., Chan K.M., Kool E.T.. ChemBioChem; 2015, 16, 1637-1646.
13. Fluorescent RNA cytosine analogue – an internal probe for detailed structure and dynamics investigations. Füchtbauer, A.F., Preus, S., Börjesson, K., McPhee, S.A., Lilley D.M.J., Wilhelmsson, L.M.. Sci. Rep.; 2017, 7, 2393.
Product Information
tC-CE Phosphoramidite (10-1516)
tCO-CE Phosphoramidite (10-1517)
- Glen Report 30.11: Introducing ribo-tCo<>/sup>
- Glen Report 30.12: DBCO-Serinol Phosphoramidite
- Glen Report 30.13: Significant Improvement of CRISPR Specificity with 2'-OMe-PACE Modifications
- Glen Report 30.14: Use of 2'-OMe-PACE Monomers During Oligo Synthesis
- Glen Report 30.15: Methylene Blue II - A Unique Dye
- Glen Report 30.16: Technical Brief – 5'-Phosphorylation of RNA
- Glen Report 30.1: Technical Snippets


