Glen Report 24.28: Technical Brief: FRETmatrix – A Method for Simulation and Analysis of FRET in Oligos
Søren Preus
Department of Chemistry
University of Copenhagen
Copenhagen, DK-2100, Denmark
Marcus Wilhelmsson
Department of Chemical and Biological Engineering/Physical Chemistry
Chalmers University of Technology
Kemivägen 10, SE-412 96 Göteborg, Sweden
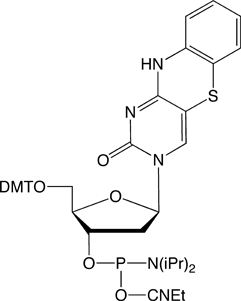
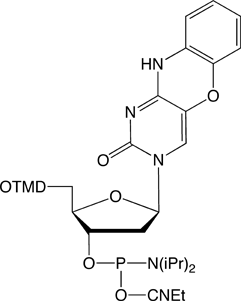
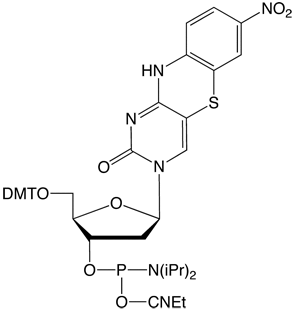
The recent introduction of nucleobase analogues tC, tCo andtCnitro (Figure 1) into the arsenal of commercial FRET probes has provided researchers with a highly useful alternative to traditional, external dyes when studying the structure, dynamics and function of nucleic acids. Since not only the interpair-distance but also the probe orientation play an important role in the energy transfer process, FRET between rigidly attached nucleobase analogues (base-base FRET) can be exploited to obtain both positional and orientational information about nucleic acid geometries without complications associated with linker flexibility. The design and interpretation of base-base FRET experiments has up until now been challenging to a large majority of researchers.
Addressing this issue, Wilhelmsson and co-workers1 have developed a freely downloadable software package (FRETmatrix) divided into two parts:
(1) the design and (2) the analysis of base-base FRET experiments.
One part of the software simulates theoretical FRET efficiencies between probes positioned in any 3D nucleic acid structure. To significantly facilitate the design and identify optimal positions of the FRET-probes in oligonucleotides of base-base FRET experiments, the software is used to predict theoretical signal changes between all possible donor-acceptor positions in a nucleic acid sequence, e.g., upon binding of a protein or some other structural change (Figure 2).
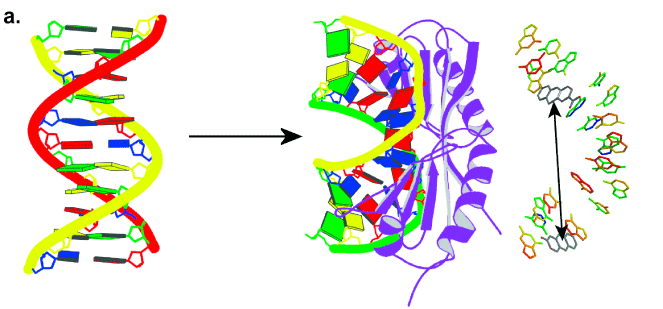

- The geometry of DNA upon binding of the TATA-binding protein (TBP)
- The predicted change in FRET efficiency at selected donor-acceptor positions.
To analyse base-base FRET experiments quantitatively, multiple time-resolved fluorescence decays can be analyzed simultaneously in the second part of the freeware. This novel, rigorous analysis may provide insight into global and/or local 3D structural features, such as the geometry of kinked DNA or the position, orientation and dynamics at specific base positions. The MATLAB-based software is freely available from the following link:
http://www.chalmers.se/chem/EN/divisions/physical-chemistry/staff/marcus-wilhelmsson/fretmatrix
Reference
- S. Preus, K. Kilsa, F.A. Miannay, B. Albinsson, and L.M. Wilhelmsson, Nucleic Acids Res, 2012.
Product Information
tC-CE Phosphoramidite (10-1516)
tC°-CE Phosphoramidite (10-1517)
tCnitro-CE Phosphoramidite (10-1518)
- Glen Report 24.21: Click DNA and RNA Ligation – A Biocompatible Triazole Linkage
- Glen Report 24.22: New Product - DBCO-dT for Copper-Free Click Chemistry
- Glen Report 24.23: Antisense Trimer Phosphoramidites - A New Method for Mutagenesis Spanning Entire Genes
- Glen Report 24.24: New Product - Alkyne-Modifier Serinol Phosphoramidite
- Glen Report 24.25: Technical Brief - Reagents for 5’-Labeling of microRNAs
- Glen Report 24.26: Advances in Copper(I)-Catalyzed Azide-Alkyne Cycloaddition (CuAAC)
- Glen Report 24.27: Introducing Oligo-Click Kits
- Glen Report 24.28: Technical Brief: FRETmatrix – A Method for Simulation and Analysis of FRET in Oligos
- Glen Report 24.29: Technical Brief: Which 5'-Amino-Modifier?
- Glen Report 24.210: Technical Brief - Improved Conditions for Deprotecting 5-Formyl-dC
- Glen Report 24.211: New Products: Universal HybridCPG™ Solid Supports

