Glen Report 24.25: Technical Brief - Reagents for 5’-Labeling of microRNAs
Introduction
MicroRNAs (miRNAs) are short, non-coding double-stranded RNAs approximately 22 nucleotides in length that are estimated to regulate 5300 human genes.1 Given their importance, several methods have been developed for the detection of miRNAs2, however, few allow the simultaneous detection of multiple miRNAs. To overcome this analytical deficiency, the Richert group has recently developed an ingenious method3 to selectively detect miRNAs on microarrays without interference from endogenous pre-mRNAs, mRNAs and other RNA species.
In the method described by Richert and Vogel3, a short oligonucleotide containing 3'-amino-dT and a 5' reporter molecule is chemically ligated to the microRNA (Figure 1) in a one-step procedure by in situ activation of the microRNA. This is specifically achieved by taking advantage of the fact that miRNAs, unlike other RNAs, are 5’-phosphorylated. The reaction is template-directed (and thus sequence specific) and can be performed together with enzymatic 3’-extension/labeling, either in solution or on a support. The short DNA labeling strand may feature one of a variety of different labels, such as a biotin group or a fluorophore.
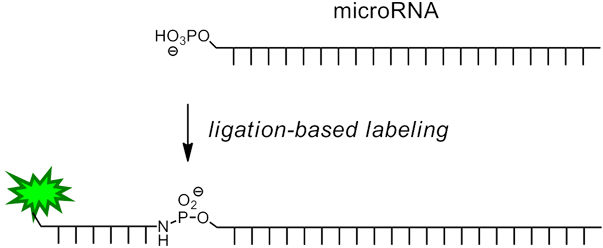
Procedure
First, a microarray is spotted with capture sequences for the miRNAs of interest. These capture sequences have attached to their 3’ terminus, a short sequence that is complementary to a labeling oligo. The labeling oligo is short – only 8 nucleotides in length – and contains a 5’ label such as biotin or a fluorescent dye, and on the 3’ terminus, a 2’,3’-dideoxy-3’-amino dT (3'-Amino-dT) nucleoside.
When the miRNAs and labeling oligo are hybridized onto the capture sequence, the 5’-phosphate of the miRNA is brought into close proximity to the 3’-amino of the labeling oligo, as shown on the left side of Figure 3. By activating the phosphate in situ with 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide hydrochloride (EDC), the 3’-amino group of the labeling oligo reacts to form a stable phosphoramidate linkage, chemically ligating the miRNA and the labeling oligo. Because the Tm of the ligated labeling oligo is significantly higher than the non-ligated labeling oligo, the unreacted labeling oligo can easily be washed away, affording a very low background signal, as seen in the pseudocolor image of the microarray in Figure 3.
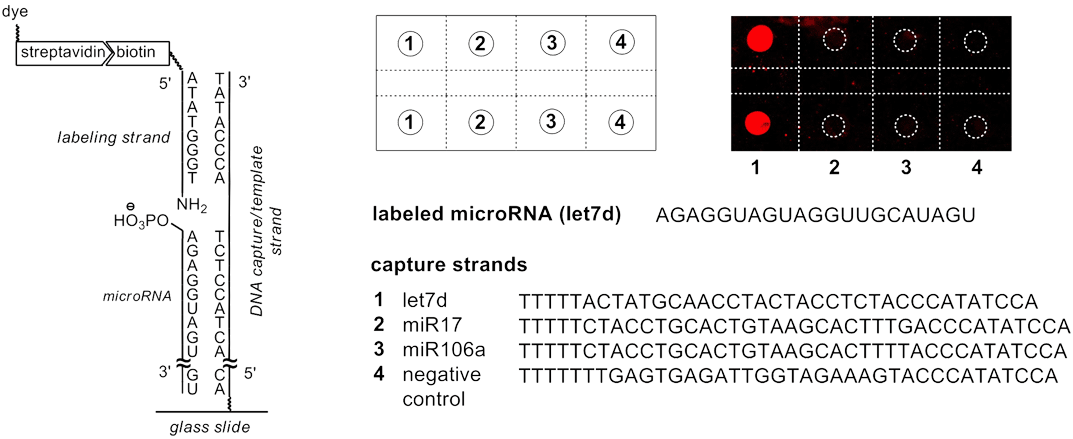
Left: Components on a molecular level. Upper Right: The spot pattern and a fluorescence image of the microarray in pseudocolor. Lower Right: The sequences of the microRNAs in 5’ to 3’-direction.
This procedure makes microarrays an attractive tool for the detection of these medically relevant species. Figure 3 details the result of a microRNA detection assay performed with a biotinylated labeling strand on a microarray. The microRNA let7d was labeled by chemical ligation with subsequent staining of the biotins via Cy5-bearing streptavidin. The fluorescence scan shows little background and exquisite3 discrimination against other RNAs, including closely related microRNAs.
For the differentiation between microRNAs that only differ in a base at the 5’-terminus, false positive signals obtained for enzymatic labeling could be avoided with chemical 5’-labeling. Chemical ligation at the 5'-terminus of the miRNA has also been successfully combined with enzymatic 3’-labeling using an on-chip protocol, with reactions performed directly on microarrays. It was found that the dual labeling with two different dyes greatly improves selectivity when detecting closely sequence-related microRNAs and can be used to massively reduce background via subtraction of unmatched signals.
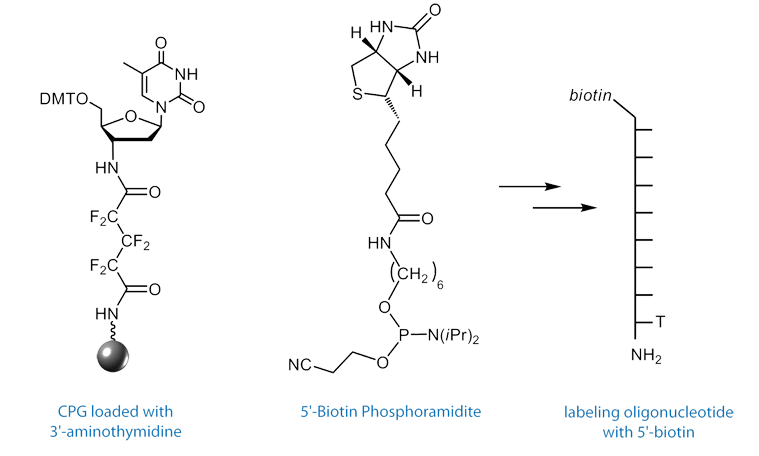
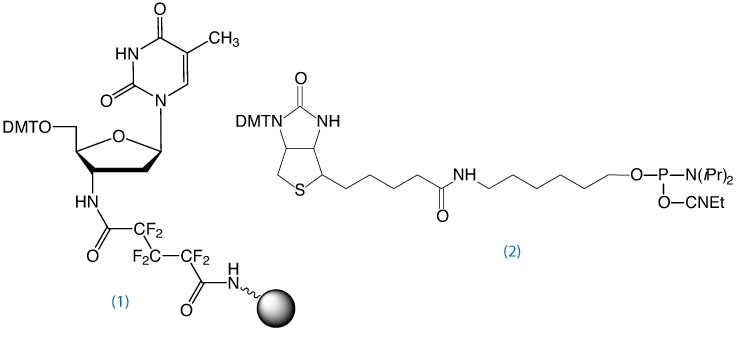
3’-Amino-dT CPG4, (1) in Figure 4, may also be used to prepare aminoterminal oligonucleotides for other applications, such as chemical primer extension.5
Oligo Synthesis and Deprotection
To obtain an oligo modified with a 3’-amino-dT residue, the Richert group developed the 3’-amino-dT CPG, (1) in Figure 4.4 After standard synthesis and labeling of the 5’ terminus with a fluorophore or biotin, the 3’-amino-dT labeled oligo is isolated on deprotection with ammonium hydroxide. Following standard purification protocols using Glen-Pak cartridges or RP HPLC, the labeling strand can be used without special steps or precautions.
Notes
This support is not compatible with modifiers requiring mild or UltraMild deprotection and it is not compatible with UltraFast deprotection with AMA.
The cleavage of the hexafluoroglutaroyl linker requires a minimum of 17 hours at 55°C with fresh ammonium hydroxide (28-30%) in water. A small percentage of the linker (~5%) may remain and full cleavage may require up to 30 hours at 55°C.
References
- B.P. Lewis, C.B. Burge, and D.P. Bartel, Cell, 2005, 120, 15-20.
- K.A. Cissell, and S.K. Deo, Anal Bioanal Chem, 2009, 394, 1109-16.
- H. Vogel, and C. Richert, ChemBioChem, 2012, 13, 1474-82.
- R. Eisenhuth, and C. Richert, Journal of Organic Chemistry, 2008, 74, 26-37.
- E. Kervio, A. Hochgesand, U.E. Steiner, and C. Richert, Proc Natl Acad Sci U S A, 2010, 107, 12074-9.
We thank Clemens Richert and Heike Vogel for their help in preparing this article. We also hope that this microarray procedure will develop into an attractive tool for the detection of these medically-relevant miRNA species.
Product Information
5'-Biotin Phosphoramidite (10-5950)
3'-Amino-dT CPG (20-2981)
- Glen Report 24.21: Click DNA and RNA Ligation – A Biocompatible Triazole Linkage
- Glen Report 24.22: New Product - DBCO-dT for Copper-Free Click Chemistry
- Glen Report 24.23: Antisense Trimer Phosphoramidites - A New Method for Mutagenesis Spanning Entire Genes
- Glen Report 24.24: New Product - Alkyne-Modifier Serinol Phosphoramidite
- Glen Report 24.25: Technical Brief - Reagents for 5’-Labeling of microRNAs
- Glen Report 24.26: Advances in Copper(I)-Catalyzed Azide-Alkyne Cycloaddition (CuAAC)
- Glen Report 24.27: Introducing Oligo-Click Kits
- Glen Report 24.28: Technical Brief: FRETmatrix – A Method for Simulation and Analysis of FRET in Oligos
- Glen Report 24.29: Technical Brief: Which 5'-Amino-Modifier?
- Glen Report 24.210: Technical Brief - Improved Conditions for Deprotecting 5-Formyl-dC
- Glen Report 24.211: New Products: Universal HybridCPG™ Solid Supports

