Glen Report 24.21: Click DNA and RNA Ligation – A Biocompatible Triazole Linkage
Afaf H. El-Sagheer and Tom Brown
School of Chemistry , University of Southampton
Highfield, Southampton SO17 1BJ, United Kingdom
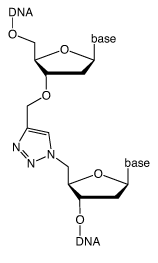
Biochemical strategies that use a combination of synthetic oligonucleotides, thermostable DNA polymerases and DNA ligases can produce large DNA constructs up to 1 megabase in length. Although these ambitious targets are feasible biochemically, comparable technologies for the chemical synthesis of long DNA strands lag far behind. The best available chemical approach is the solid-phase phosphoramidite method, which can be used to assemble DNA strands up to 150 bases in length. Beyond this point deficiencies in the chemistry make it difficult to produce pure DNA.
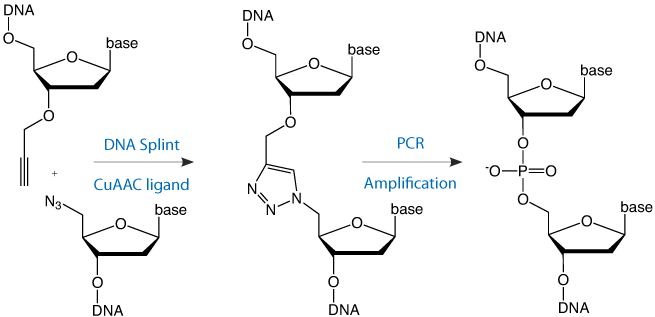
A possible alternative approach to the chemical synthesis of large DNA strands is to join together synthetic oligonucleotides by chemical methods. Recently click ligation by the copper-catalyzed azide-alkyne (CuAAC) reaction has been shown to facilitate this process, and a biocompatible triazole linkage has been developed that mimics a normal phosphodiester group.1
This requires an oligonucleotide with a 5’-azide and another with a 3’-propargyl group. The two oligonucleotides can be joined together by splint mediated ligation to produce a triazole linkage at the ligation site (Figure 1, Page 2). Three or more oligonucleotides can be joined together by this methodology using internal difunctionalized oligonucleotides with 5’-azide and 3’-propargyl groups in the same strand. The alkyne and azide oligonucleotide strands can be prepared by standard protocols and, as it is chemical rather than enzymatic, the chemical ligation reaction is compatible with a wide range of other oligonucleotide modifications.
Click ligation has been employed to synthesize DNA constructs up to 300 bases in length and, when the resulting triazole linkage is placed in a PCR template, various DNA polymerases correctly copy the entire base sequence.1 In vitro transcription through the modified linkage has also been successfully demonstrated.2
Cyclic DNA duplexes with potential therapeutic applications can be made using this methodology and have been shown to be substrates for rolling circle amplification.1
This modified triazole linkage has shown in vivo biocompatibility; an antibiotic resistance gene containing triazole linkages is functional in E. coli1 and a triazole-containing gene for a (non-essential) fluorescent protein has been expressed.3
A recent NMR study of a DNA duplex containing the triazole linkage has provided a rationale to explain the biocompatibility of this linkage.4
Click ligation in the RNA field has enabled the synthesis of an enzymatically active hammerhead ribozyme with the triazole linkage located at the substrate cleavage site.5
Possible applications of click ligation include assembly of long DNA or RNA strands incorporating unusual sugar or backbone modifications (including chimeric molecules), epigenetic modifications, fluorescent dyes and other reporter groups which might be unstable to enzymatic ligation conditions. In the RNA world, segmental labeling followed by the assembly of large chemically-modified RNA constructs for functional studies is a distinct possibility.
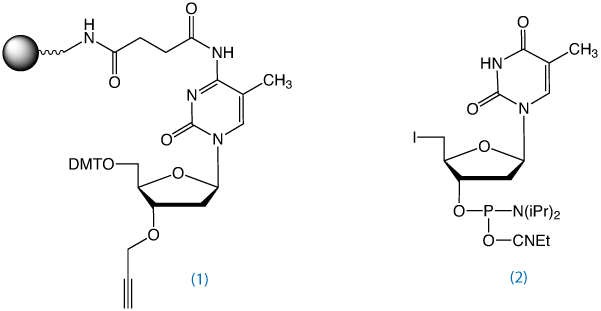
3'-Propargyl-5-Me-dC-CPG, (1) in Figure 2, is used to prepare 3'-propargyl modified oligonucleotides. In collaboration with Tom Brown's group at the University of Southampton, Glen Research has added this product to the catalog of reagents for Click Chemistry.
References
- A.H. El-Sagheer, A.P. Sanzone, R. Gao, A. Tavassoli, and T. Brown, Proc Natl Acad Sci U S A, 2011, 108, 11338-43.
- A.H. el-Sagheer, and T. Brown, Chem Commun (Camb), 2011, 47, 12057-8.
- A.P. Sanzone, A.H. El-Sagheer, T. Brown, and A. Tavassoli, Nucleic Acids Res, 2012.
- A. Dallmann, et al., Chemistry, 2011, 17, 14714-7.
- A.H. El-Sagheer, and T. Brown, Proc Natl Acad Sci U S A, 2010, 107, 15329-34.
Protocols
Conversion of 5’-dC or 5’-dT to 5’-azido dC or 5’-azido dT for oligonucleotides with an unmodified 3’-end
Oligonucleotides were assembled on the 0.2 or 1.0 μmole scale (trityl-off) with a normal 5’-HO-dC or 5'-HO-dT terminus. To convert the 5'-hydroxyl group to 5'-iodo,1 the protected oligonucleotide attached to the synthesis column was treated with a 0.5 M solution of methyltriphenoxyphosphonium iodide in DMF (1 mL), which was periodically passed through the column via two 1 mL syringes over 15 min at room temperature. The column was then washed several times with dry DMF. To convert the 5'-iodo to 5'-azido, sodium azide (30 mg) was suspended in dry DMF (1 mL), heated for 10 min at 70 °C then cooled down. The supernatant was taken up into a 1 mL syringe, passed back and forth through the column then left at room temperature overnight (or for 5 hr at 55 °C with periodic mixing). The column was then washed with DMF, acetonitrile and dried by the passage of a stream of argon gas. The resultant 5'-azide oligonucleotide was cleaved from the solid support and deprotected by heating in aqueous ammonia for 5 hr at 55 °C.
5'-Azido oligonucleotides can also be synthesized using the 5'-iodo-dT monomer, (2) in Figure 2, and converting it to 5'-Azido using sodium azide in DMF as described above.
Conversion of 5’-dC or 5’-dT to 5’-azido dC or 5’-azido dT for oligonucleotides with 3’-propargyl-5-Me-dC at the 3’-end
5'-O-(4,4'-Dimethoxytrityl)-3'-O-propargyl-5-methyl-2'-deoxyCytidine on a solid support was packed into a TWIST column and used to assemble the required sequence in the 3'- to 5'-direction (standard phosphoramidite oligonucleotide synthesis) with 5'-iodo dT, 5'-HO-dT or 5'-HO-dC at the 5'-end. The 5'-hydroxyl or iodo groups were then converted to azide using the conditions described above for the synthesis of the 5'-azide oligonucleotides using unmodified solid support.
In the case of the 3’-propargyl-5-Me-dC functionalized resin, the linkage is not very stable to prolonged contact with the azide solution in DMF, therefore some of the oligonucleotide will be released from the resin into solution. To achieve better yield, the DMF solution and washings were evaporated, the residue was dissolved in water and desalted using NAP-25 columns. After evaporating the water under vacuum, the residue can be combined with the washed and dried resin and heated in aqueous ammonia for 5 hr at 55 °C for deprotection.
Reference
- 1. G.P. Miller, and E.T. Kool, J Org Chem, 2004, 69, 2404-2410.
Product Information
5'-I-dT-CE Phosphoramidite (10-1931)
3'-Propargyl-5-Me-dC CPG (20-2982)
- Glen Report 24.21: Click DNA and RNA Ligation – A Biocompatible Triazole Linkage
- Glen Report 24.22: New Product - DBCO-dT for Copper-Free Click Chemistry
- Glen Report 24.23: Antisense Trimer Phosphoramidites - A New Method for Mutagenesis Spanning Entire Genes
- Glen Report 24.24: New Product - Alkyne-Modifier Serinol Phosphoramidite
- Glen Report 24.25: Technical Brief - Reagents for 5’-Labeling of microRNAs
- Glen Report 24.26: Advances in Copper(I)-Catalyzed Azide-Alkyne Cycloaddition (CuAAC)
- Glen Report 24.27: Introducing Oligo-Click Kits
- Glen Report 24.28: Technical Brief: FRETmatrix – A Method for Simulation and Analysis of FRET in Oligos
- Glen Report 24.29: Technical Brief: Which 5'-Amino-Modifier?
- Glen Report 24.210: Technical Brief - Improved Conditions for Deprotecting 5-Formyl-dC
- Glen Report 24.211: New Products: Universal HybridCPG™ Solid Supports

