Glen Report 20.28: Site-specific incorporation of functional components into RNA by transcription using unnatural base pair systems
Introduction
RNA-based research and biotechnology are growing rapidly and extensively. In the process, researchers have long sought a method for allowing the site-specific incorporation of extra components, with a functional group of interest, into desired positions of RNA molecules. Such a method could be provided by creating extra, unnatural base pairs to augment the natural A-T and G-C pairs of DNA, and to expand the genetic alphabet1-4. Hirao’s group has now developed the unnatural base pairs between 7-(2-thienyl)-imidazo[4,5-b]pyridine (Ds) and pyrrole-2-carbaldehyde (Pa)5, and 2-amino-6-thienylpurine (s) and Pa6, which can be utilized in transcription for the site-specific, enzymatic incorporation of functional components into RNA by T7 RNA polymerase. The Ds-Pa pair complementarily mediates the incorporation of the triphosphate substrates of Ds (DsTP) and Pa (PaTP) into RNA by T7 transcription. A series of modified Pa bases, bearing functional groups attached to position 4 of Pa via an aminopropynyl linker, is also incorporated into RNA opposite Ds in DNA templates. Furthermore, Pa can be used as a template base for the enzymatic incorporation of the fluorescent s base, as a triphosphate substrate (sTP), into RNA. Glen Research has already begun offering the amidites of dDs and dPa for DNA template synthesis. (See Figure 1 and Glen Report, Vol. 20, No. 1 in 2008). We are now supplying Biotin PaTP and sTP for their incorporation by T7 transcription (Figure 1).
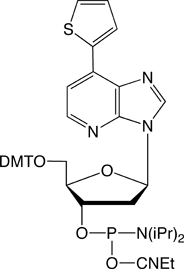
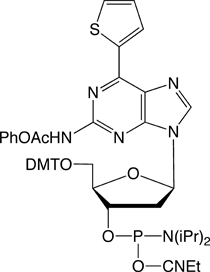
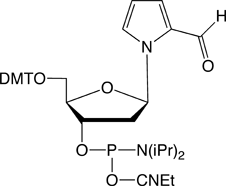
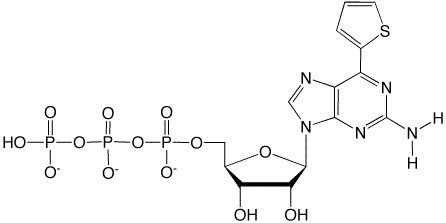

Biotin PaTP
Biotinylated RNA molecules are routinely used for immobilization on avidin supports or for chemiluminescent detection, using streptavidin coupled to alkaline phosphatase. Biotin PaTP can be site-specifically incorporated into RNA, opposite dDs at a desired position in DNA templates, by T7 transcription5. Thus, this method facilitates the immobilization and detection of a target RNA without the loss of the intrinsic activity of the RNA molecule.
sTP
The s base is strongly fluorescent, absorbing at 299 and 352 nm and emitting at 434 nm, with a quantum yield of 0.41 at pH 7.07. The fluorescent intensity of s in DNA and RNA molecules changes according to the structural environment, and is quenched depending on the degree of stacking interactions with adjacent bases6,7. Since the stacking manner in RNA molecules directly reflects their tertiary structure, site-specific s labeling is useful for studying the dynamics of the local structural features and changes of RNA molecules. The fluorescent s base can be site-specifically incorporated into RNA opposite dPa in DNA templates, by T7 transcription.
Applications of non-natural nucleosides
Local Structural Analysis of DNA
- Use ds Amidite
- Introduce s base in single stranded DNA
- Fluorescence intensity of ds decreases when ds-containing oligo hybridizes
- Fluorescence ds probing is useful for analyzing oligonucleotide behavior in many practical applications
- FRET experiments possible
- Tetrahedron, 2007, 63, 3528-3537.
Local Structural Analysis of RNA
- sTP / dPa system
- Make DNA Template containing dPa using amidite
- Produce fluorescent RNA using sTP during transcription
- Fluorescence intensity of s decreases when s-containing RNA hybridizes or forms structures, allowing local structure analysis
- Nucleic Acids Res., 2007, 35, 5360-5369.
Site-specific Biotinylation of RNA
- The dDs / Biotin PaTP system
- PCR amplify a target RNA using a pair of primers - one primer introduces T7 promoter and the other contains dDs analogue
- PCR amplification produces a DNA template with dDs
- T7 transcription using Biotin PaTP produces RNA containing a Biotin at the specific position of Ds
- Nature Methods, 2006, 3, 729-735.

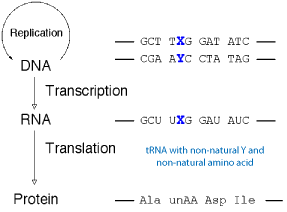
With an unnatural base pair [X-Y] functioning in replication, transcription and translation, it becomes possible to incorporate nucleotide analogs at specific sites into RNA and introduce amino acid analogues (unAA) into proteins.
Science, 1989, 244, 182-188.
Nucleic Acids Res., 2002, 30, 4692-4699.
References
- E.T. Kool, Curr. Opin. Chem. Biol., 2000, 4, 602-608.
- A.A. Henry, and F.E. Romesberg, Curr. Opin. Chem. Biol., 2003, 7, 727-733.
- S.A. Benner, and A.M. Sismour, Nat. Rev. Genet., 2005, 6, 533-543.
- I. Hirao, Curr. Opin. Chem. Biol., 2006, 10, 622-627.
- I. Hirao, et al., Nat. Methods, 2006, 3, 729-735.
- M. Kimoto, et al., Nucleic Acids Res., 2007, 35, 5360-5369.
- T. Mitsui, M. Kimoto, R. Kawai, S. Yokoyama, and I. Hirao, Tetrahedron, 2007, 63, 3528-3537.
Intellectual Property
This product is covered by patents or patents pending owned by TagCyx Biotechnologies. Purchase of this product includes a limited license to use this product solely for research. This license specifically excludes: (a) therapeutic or diagnostic applications (including products or services that incorporate this product), (b) any in vivo toxicity/safety study in support of an investigational new drug application (or foreign counterpart), (c) resale, or (d) gene functionalization activities (including products or services that incorporate data derived from gene functionalization activities) if such activities have commercial application. All of the above require a separate license from TagCyx Biotechnologies. Neither this product nor any product created through its use may be used in human clinical trials.
Product Information
All products in this article have been discontinued.
- Glen Report 20.21: Phosphonoacetate (PACE) Oligonucleotides Introduction
- Glen Report 20.22: Synthesis, Cleavage and Deprotection of PACE Oligonucleotides
- Glen Report 20.23: Novel Reagents for Modification and Labelling
- Glen Report 20.24: Deprotection - Volume 1 - Deprotect to Completion
- Glen Report 20.25: New Universal Support - Glen UnySupport
- Glen Report 20.26: New Products for Attachment of Oligonucleotides on Gold Surfaces
- Glen Report 20.27: 5'-Dichloro-Dimethoxy-Fluorescein (JOE™) Phosphoramidite
- Glen Report 20.28: Site-specific incorporation of functional components into RNA by transcription using unnatural base pair systems

