Glen Report 20.24: Deprotection - Volume 1 - Deprotect to Completion
Introduction
This article offers practical information that will help newcomers to the field of oligo synthesis to understand the various considerations before choosing the optimal deprotection strategy, as well as the variety of options that are available for deprotection. It is not the intent of these articles to provide a comprehensive, fully referenced review of deprotection strategies in oligonucleotide synthesis - they are simply guidelines. For more detailed information, see, for example, the review1 by Beaucage and Iyer. This is the first of a series of articles on deprotection that will be posted on our web site.
Oligo deprotection can be visualized in three parts: cleavage, phosphate deprotection, and base deprotection. Cleavage is removal from the support. Phosphate deprotection is the removal of the cyanoethyl protecting groups from the phosphate backbone. Base deprotection is the removal of the protecting groups on the bases or modifier. There are many considerations when approaching oligo deprotection, as shown in the Box on the right. However, when reviewing the procedures available to deprotect any oligonucleotide, you must heed the primary consideration: First, Do No Harm. You can then proceed with confidence to Deprotect to Completion.
First, Do No Harm!
Determination of the appropriate deprotection scheme should start with a review of the components of the oligonucleotide to determine if any group is sensitive to base and requires a mild deprotection or if there are any pretreatment requirements. Sensitive components are usually expensive components so it is imperative to follow the procedure we recommend for any individual component. For example, the presence of a dye like TAMRA or HEX will require a different procedure from regular unmodified oligonucleotides. Similarly, an oligo containing a base-labile monomer like 5,6-dihydro-dT will have to be treated according to the procedure that is noted on the product insert (Analytical Report, Certificate of Analysis, or Technical Bulletin). Occasionally, some products require a special pretreatment to prevent unwanted side reactions. For example, amino modifiers use a special diethylamine pretreatment to improve the overall yield of the amino-labelled oligo. If the oligo has several unusual components, you must follow the mildest procedure recommended and, yes, things can get complicated fast. Volume 2 will focus on this complex topic.
RNA deprotection is unique because of the necessity to retain the 2' protecting group during cleavage and base deprotection. 2'-OMe-RNA and 2'-F-RNA, however, are virtually identical to DNA during deprotection. But, if a hybrid oligonucleotide contains even a single RNA linkage (with the exception of a 3'-ribonucleoside linkage), the oligo must be treated as RNA. See the appropriate RNA deprotection protocols:
PROCEDURE FOR THE SYNTHESIS, DEPROTECTION AND ISOLATION OF RNA USING TOM-PROTECTED MONOMERS
Another consideration for potential harm is loss of trityl group during vacuum concentration of the oligo solution prior to purification, which will reduce product yield. During evaporation the heat should be turned off the vacuum concentrator to avoid loss of the DMT group. It should be noted that most DMT-on purification protocols, including Poly-Pak™ and Glen-Pak™, do not require evaporation of the deprotection solution prior to purification.
A unique case for potential harm is an oligonucleotide containing a 5'-amine protected with the MMT protecting group (e.g., 10-1906). In this situation, deprotection should not be carried out at > 37°C to avoid thermal loss of the MMT group.
Cleavage
On classic synthesizers from Applied Biosystems, the cleavage of the oligo from the synthesis support can be carried out separately on the machine, prior to deprotection. As a result, many researchers still carry out the cleavage reaction separately and so the time required to do this is mentioned at the beginning of each Deprotection section. However, most researchers do a one step cleavage/deprotection reaction, which has the advantage of ensuring optimal yields. The only downside to this strategy is the fact that the basic solution at elevated temperatures will dissolve a small amount of silica from CPG and a white insoluble residue will be apparent if the deprotection solution is evaporated to dryness. However, any residual silicate is easily removed by filtration, desalting or any purification procedure.
Deprotect to Completion
Volume 1: Deprotect to Completion
- Do I have very special components in my oligo or not?
- Am I in a rush or not?
- Do I have one or many oligos to treat?
- Do I need/want to purify my oligo after deprotection or not?
- Does my oligo contain RNA, 2’-OMe-RNA, or 2’-F-RNA linkages?
The rate-determining step in oligonucleotide synthesis is more than likely the removal of the protecting group on the G base. Ignore this at your peril since, traditionally, one of the most common reasons for poor performance of oligonucleotides is the presence of a small percentage of the G protecting groups remaining in the final product oligonucleotide. Chromatographic methods may miss the presence of the G protecting groups but these are readily revealed by mass spectral analysis. What are the options with attendant pros and cons for oligonucleotide deprotection?
Regular Deprotection
The cleavage reaction with concentrated ammonium hydroxide (28 to 33% NH3 in water), if carried out separately, is normally considered to be 1 hour at room temperature. Deprotection using ammonium hydroxide is the most traditional method and dates back to the earliest days of oligonucleotide synthesis. One of the critical issues when using ammonium hydroxide, which is water saturated with ammonia gas, is to keep the solution fresh. We aliquot and store ammonium hydroxide in the refrigerator in portions appropriate for use in 1 week. Using an old bottle of ammonium hydroxide is false economy since the resulting oligos are not going to be completely deprotected.
| dG Protection | Temperature | Time |
|---|---|---|
| iBu-dG | RT | 36h |
| 55°C | 16h | |
| 65°C | 8h | |
| dmf-dG , Ac-dG | RT | 16h |
| 55°C | 4h | |
| 65°C | 2h | |
| iPr-Pac-dG | RT | 2h |
| 55°C | 0.5h | |
| Table 1, Page 8 shows the various times and temperatures appropriate for deprotection with FRESH ammonium hydroxide. | ||
UltraFAST Deprotection
Using the UltraFAST procedure, cleavage of the oligonucleotide from the support is performed using AMA2 which is a 1:1 mixture (v/v) of aqueous Ammonium hydroxide and aqueous MethylAmine. If carried out separately, it is accomplished in 5 minutes at room temperature.
UltraFAST deprotection allows 5-10 minute deprotection of oligonucleotides using AMA. It is important to note that the UltraFAST system requires acetyl (Ac) protected dC to avoid base modification at the C base if Bz-dC is used. The three other monomers remain unchanged and the system works equally well with iBu-, Ac-, or dmf-dG, the last being our preferred dG phosphoramidite.
The deprotection step is carried out at 65°C for 5 minutes. Deprotection can also be carried out at lower temperatures as shown in Table 2. In all cases, no base modification has been observed.
Figure 1 illustrates the differences in RP HPLC between partially and fully deprotected oligos, DMT-off and DMT-on.
| dG Protection | Temperature | Time |
|---|---|---|
| iBu-dG, dmf-dG or Ac-dG | RT | 120 min. |
| 37°C | 30 min. | |
| 55°C | 10 min. | |
| 65°C | 5 min. | |
| Note: UltraFAST system requires acetyl (Ac) protected dC to avoid base modification at the C base. | ||
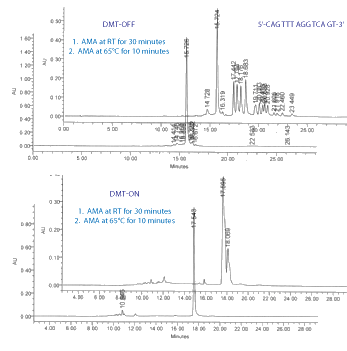
RP HPLC Conditions Column: Waters X-Bridge C18, 250 x 4mm; Buffers: A - ACN; B - 0.1M TEAA , pH 7. DMT-off Gradient: 3-15% B over 15 min. DMT-on Gradient: 3-40% B over 15 min. Flow rate: 1mL/min.
UltraMild Deprotection
Cleavage is not carried out separately when using UltraMILD techniques. Since many of our nucleosides and dye products are not stable to deprotection with ammonium hydroxide or AMA, the procedure to deprotect the labelled oligonucleotide must be changed.
We often recommend using the UltraMILD monomers (Pac-dA, Ac-dC and iPr- Pac-dG) and deprotection with potassium carbonate in methanol. In this way, some of these very sensitive oligonucleotides can be conveniently isolated. If capping is carried out using Cap A containing phenoxyacetic anhydride, it is possible to deprotect UltraMILD oligonucleotides in 4 hours at RT with 0.05M potassium carbonate in methanol or 2 hours at RT with ammonium hydroxide. Alternatively, using the regular Cap A containing acetic anhydride, it is necessary to deprotect overnight at room temperature to remove any Ac-dG formed during the capping step. For TAMRA containing oligonucleotides, an alternative deprotection3 may be carried out using t-butylamine/methanol/water (1:1:2) overnight at 55°C. Another option that we have found to be excellent uses t-butylamine/water (1:3) for 6 hours at 60°C. In this case, the regular protecting groups on the monomers may be used. An even milder approach has been described as “Ultra-UltraMild".4 In this technique, Q-supports5 are combined with UltraMild monomers to allow extremely gentle deprotection. After completion of the synthesis, the solid support is dried and treated overnight at 55°C with a solution containing 10% (v/v) diisopropylamine (iPr2NH) in 0.25 M ß-mercaptoethanol in MeOH.
Summary
Successful oligonucleotide cleavage and deprotection require consideration of the deprotection conditions for each product and some products may require pretreatment or special deprotection conditions. Each synthesis should be reviewed to ensure the products have compatible deprotection conditions. Special deprotection requirements can be found on our Analytical Reports, Certificates of Analysis, Technical Bulletins, and Website: http://www.glenresearch.com.
References
- S.L. Beaucage and R.P. Iyer, Tetrahedron, 1992, 48, 2223-2311.
- M.P. Reddy, N.B. Hanna, and F. Farooqui, Nucleos Nucleot, 1997, 16, 1589-1598.
- B. Mullah and A. Andrus, Tetrahedron Lett, 1997, 38, 5751-5754.
- L.C.J. Gillet, J. Alzeer, and O.D. Scharer, Nucleic Acid Res, 2005, 33, 1961-1969.
- R.T. Pon, and S.Y. Yu, Nucleic Acids Res, 1997, 25, 3629-3635.
- Glen Report 20.21: Phosphonoacetate (PACE) Oligonucleotides Introduction
- Glen Report 20.22: Synthesis, Cleavage and Deprotection of PACE Oligonucleotides
- Glen Report 20.23: Novel Reagents for Modification and Labelling
- Glen Report 20.24: Deprotection - Volume 1 - Deprotect to Completion
- Glen Report 20.25: New Universal Support - Glen UnySupport
- Glen Report 20.26: New Products for Attachment of Oligonucleotides on Gold Surfaces
- Glen Report 20.27: 5'-Dichloro-Dimethoxy-Fluorescein (JOE™) Phosphoramidite
- Glen Report 20.28: Site-specific incorporation of functional components into RNA by transcription using unnatural base pair systems

