Glen Report 14.11: Novel Universal Support Features Rapid Amide-Assisted Dephosphorylation
Universal Support II
- Fast - cleavage and dephosphorylation in 20 minutes at room temperature
- Mild - cleavage reagent is 2M ammonia in methanol
- Compatible with UltraMild, normal, and UltraFast deprotection
- Cost-effective - comparable in price to regular 2'-deoxynucleoside supports
The vast majority of oligonucleotide syntheses are carried out on supports to which the first nucleoside has been pre-attached. Following routine cleavage and deprotection, this nucleoside becomes the terminal nucleoside of the target oligonucleotide. For standard DNA and RNA synthesis, this requires an inventory of four deoxynucleoside and four ribonucleoside supports. A universal support (a support without the first nucleoside attached) offers the following advantages:
- eliminates the need for an inventory of nucleoside supports.
- minimizes the possibility of error in the selection of the correct support type.
- reduces time and eliminates the possible error in the generation of an array of nucleoside supports in 96 well synthesizers.
- allows the preparation of oligonucleotides containing a 3'-terminal nucleoside which is not available as a support.
Since these are major benefits, why is the use of a universal support not, well, universal? Indeed, universal-type supports are standard in the synthesis of amino-, thiol-, and other modified oligonucleotides. The major hurdle to overcome is to find conditions to eliminate the terminal phosphate, produced from the first nucleoside phosphoramidite addition cycle, to the required terminal hydroxyl group. Let's look at the problem in detail.
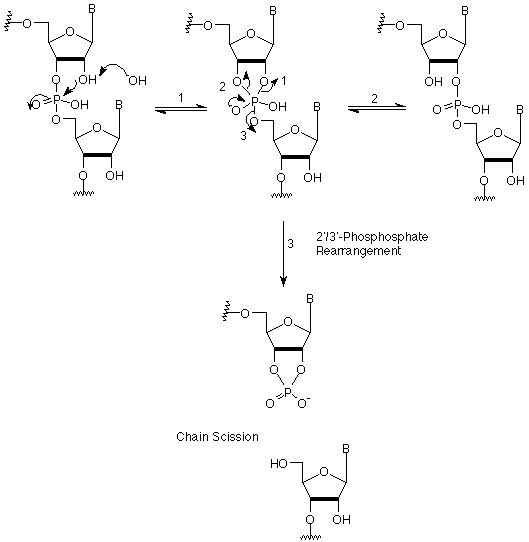
Universal supports are based on the ribonucleoside elimination model exemplified by our Universal Support (1) (Figure 1), originally introduced in 1997 and known as the McLean support.1 The instability of RNA to strongly basic conditions is caused by the proximity of the 2'-OH group to the phosphodiester group. Attack of the 2'-OH on the adjacent phosphorus gives rise to an energetically favorable 5-membered transition state, which can open up again to form a mixture of 2'- and 3'-phosphodiester internucleotide linkages, or can lead to chain scission by elimination of the 3'- or 5'-hydroxyl group (Scheme 1). In the case of our Universal Support (1), cleavage from the support by hydrolysis of a succinate or, better, the hydroquinone-O,O'-diacetate (Q) linkage, generates a hydroxyl group adjacent to the terminal phosphodiester linkage. Additional base treatment leads to the elimination of the terminal phosphate group and formation of the desired 3'-OH (Scheme 2). A similar strategy using a neighboring hydroxyl group to facilitate elimination was described by Wengel and coworkers.2 A neat variation by Lyttle3 and the group at Biosearch Technologies uses a linkage to a polymeric support that is not hydrolyzed by base, so that extended base treatment releases the dephosphorylated oligo, while leaving any undesired by-product still attached to the support.
The main impediment to the universal adoption of a universal support has been the aggressively basic conditions required to complete the elimination reaction to release the terminal hydroxyl group. The standard reagents used in oligonucleotide deprotection are ammonium hydroxide and aqueous methylamine, which are popular since they are completely volatile. Using these reagents to carry out the elimination reaction requires either high temperature, with attendant high pressure, or extended reaction times. The situation can be improved by adding metal ions to the mix, and Li+, Na+ and Zn2+ have all been used to speed up the elimination reaction, presumably by stabilizing the 5-membered transition state. However, the speed and simplicity of evaporation of the deprotection solution to give the crude oligonucleotide without desalting is not possible with these ionic additives.
Considerably improved universal supports, using neighboring aminomethyl or diamino-ethyl groups to assist the elimination reaction, were recently described by Azhayev.4 Using volatile ammonium hydroxide or aqueous methylamine, terminal dephosphorylation was significantly speeded up and even could be achieved using aqueous zinc chloride. We felt that this setup offered genuine advantages, especially if the oligonucleotides contained base-labile components, but was not quite ideal for mainstream applications.
Azhayev's group then went a step further in the investigation of neighboring group assistance in the dephosphorylation reaction. Amide groups may be considered to be weak N-H acids and can display basic properties in ammonium hydroxide or aqueous methylamine. (±)-3-Amino-1,2-propanediol was acylated to form several amide structures followed by dimethoxytritylation at the 1-position. The 2-hydroxyl was used to attach to supports via succinate or Q linkages and oligonucleotide synthesis was carried out as normal. Hydrolysis and dephosphorylation in a variety of basic solutions was investigated using T6 as the target oligo. Yields of 75-88% ( relative to T6 prepared normally) were achieved using the Q-support, trifluoroacetamide and 2-9M ammonia in methanol-toluene or methanol-dioxane. Interestingly, aqueous ammonium hydroxide led to only 43% yield of T6 with the remainder being the side product formed by ß-elimination of the cyanoethyl phosphate protecting group rather than the desired 3'-OH (Scheme 3).
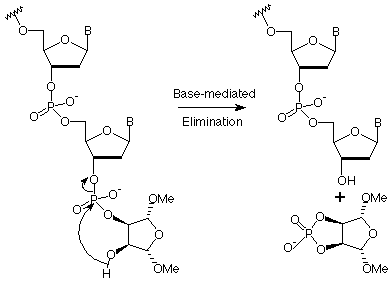
It was observed that the Q-linker was far superior to the equivalent succinate, indicating that faster generation of the 2-OH, relative to the b-elimination of the cyanoethyl phosphate protecting group is essential for successful dephosphorylation. Moreover, it was apparent that the acyl group on the amine had very little effect on the dephosphorylation reaction. Azhayev reasoned that using a succinate linker to attach the 3-amino group to the support and protecting the 2-OH with a labile protecting group should set up the identical amide assisted elimination in mildly basic conditions on the solid support. In this way, the dephosphorylation reaction would eliminate the desired 3'-OH oligonucleotide into solution and the product of any b-elimination competing side reaction would remain bound to the support (Scheme 4).
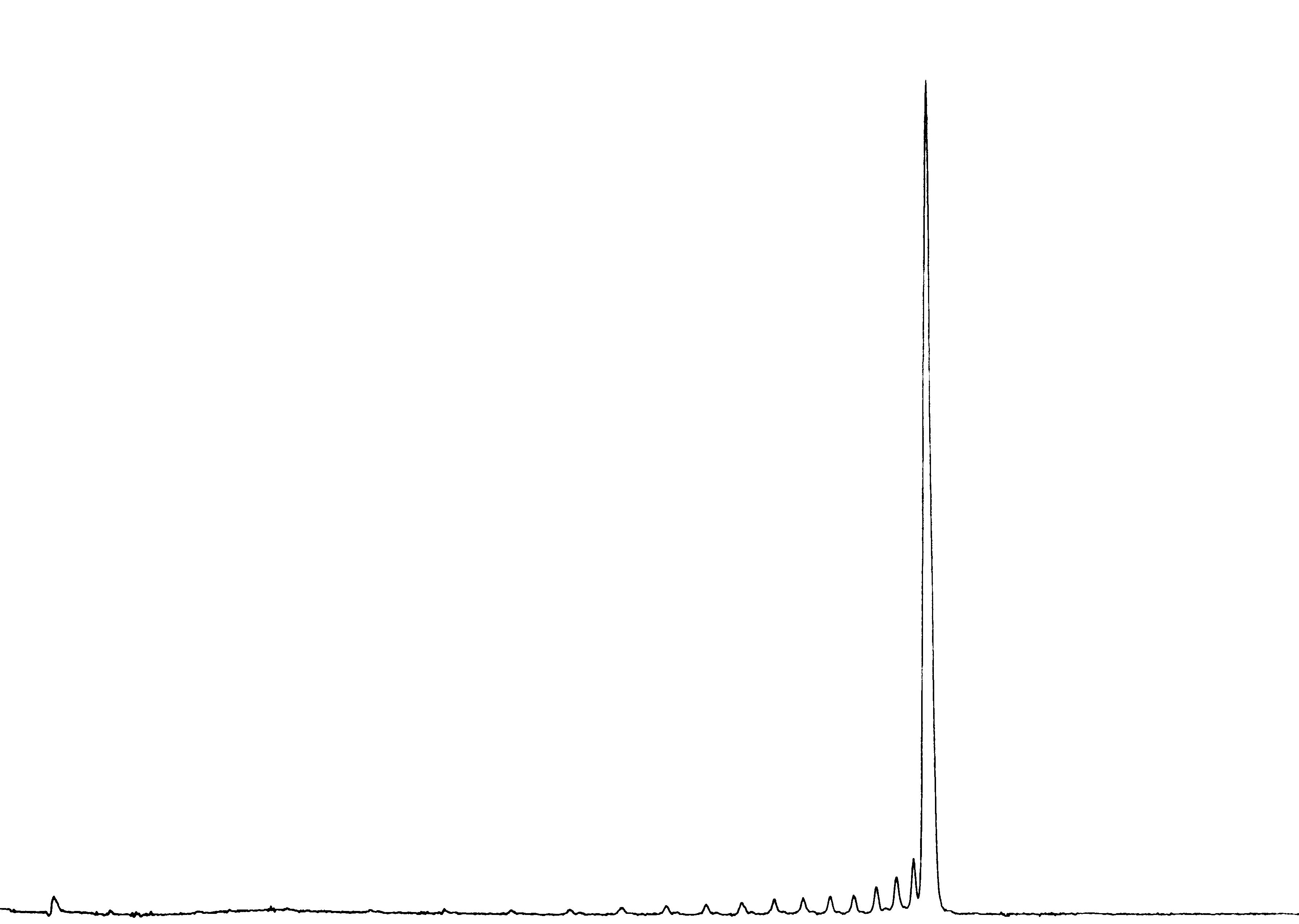
A variety of 2-O esters were tested to determine the optimal protecting group in conjunction with various mild hydrolysis conditions. The combination of 2-O formyl ester, Universal Support II (2, R=H)(Figure 1, Page 4), and hydrolysis with commercially available 2M ammonia in methanol for 1 hour at room temperature led to the optimal yield (>80%) and equivalent purity compared to T6 prepared normally5, as shown in Figures 2 and 3.
Once the cleavage and dephosphorylation reactions are complete, the deprotection of the standard bases can be achieved as follows:
- UltraMild &endash; add potassium carbonate in methanol or ammonium hydroxide and leave for a further 4 hours at room temperature.
- Regular &endash; add ammonium hydroxide and incubate for the normal time and temperature.
- UltraFast &endash; add AMA and deprotect as normal.
Universal Support II has been shown to generate oligonucleotides with the same efficacy in polymerase extension reactions as regular oligos. Despite the mild elimination reaction, oligonucleotides up to 50mer in length can be prepared routinely without substantial loss of oligo during the synthesis cycles.
References
- S. Scott, P. Hardy, R.C. Sheppard, and M.J. McLean, Innovations and Perspectives in Solid Phase Synthesis, 3rd International Symposium, 1994, 115-124.
- C. Scheuer Larsen, C. Rosenbohm, T.J.D. Jorgensen, and J. Wengel, Nucleosides & Nucleotides, 1997, 16, 67-80.
- M.H. Lyttle, D.J. Dick, D. Hudson, and R.M. Cook, Nucleosides & Nucleotides, 1999, 18, 1809-1824.
- A.V. Azhayev, Tetrahedron, 1999, 55, 787-800.
- A.V. Azhayev and M. Antopolsky, Personal Communication. Patent Pending.
Product Information
Universal Support and Universal Support II have been discontinued. Please see Universal Support III
- Glen Report 14.11: Novel Universal Support Features Rapid Amide-Assisted Dephosphorylation
- Glen Report 14.12: A New Simplified 3'-Amino-Modifier CPG
- Glen Report 14.13: Preparation of Oligonucleotides Containing Abasic Sites
- Glen Report 14.14: PC Biotin and Related Photocleavable Modifiers
- Glen Report 14.15: New OPeC™ Reagents for Synthesis by Native Ligation of Oligonucleotide-Peptide Conjugates
- Glen Report 14.16: Product Update - How Are Methyl Triester Linkages Prepared?


 (1) Universal Support
(1) Universal Support 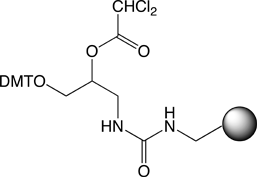 (2) Universal Support II
(2) Universal Support II