Glen Report 30.14: Use of 2'-OMe-PACE Monomers During Oligo Synthesis
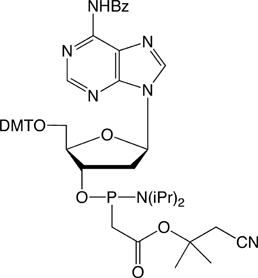
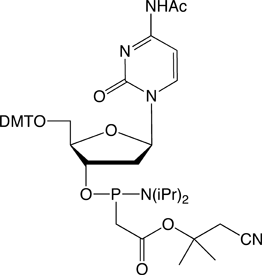
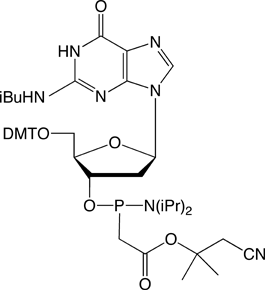
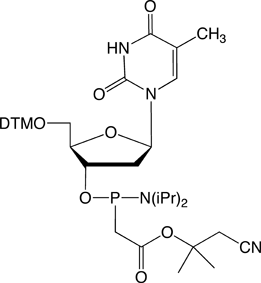
The structures of the DNA and 2'-OMe-RNA PACE monomers are shown in Figure 3.
Optimal Cycle
We recommend using DCI as an activator (30-3150-XX) and a 15 minute coupling time.
As with methyl phosphonates, the 2'-OMe-PACE modification is degraded by N-methylimidazole during capping and is susceptible to cleavage during aqueous oxidation using iodine. For this reason, we recommend using Unicap (40-4410-XX), a phosphoramidite-based capping reagent, and 0.5 M CSO (40-4632-XX), a non-aqueous oxidizer, for best results.
Following coupling of the 2'-OMe PACE monomer, cap using Unicap with a regular coupling time and then oxidize using the 0.5 M CSO for 3 minutes.
Alternative Cycle
Alternatively, a 33 minute coupling time using 0.45 M tetrazole, oxidation using low-water iodine (40-4032-XX) followed by capping with 6.5% DMAP as Cap B will give acceptable results.
Deprotection
Pre-treat the synthesis column with 1.5% DBU in anhydrous acetonitrile for 60 minutes at room temperature to remove 1,1-dimethyl-2-cyanoethyl protecting groups. Rinse the column with ACN, dry under with argon and complete deprotection with 40% Methylamine for 2 hours at room temperature.
Purification and Analysis of 2'-OMe-PACE oligos
Using the optimized synthesis conditions listed above, a simple oligo was synthesized for ease and accuracy of analysis:
5'-TXT TXT TXT TTT-3', where X is 2'-OMe U-PACE.
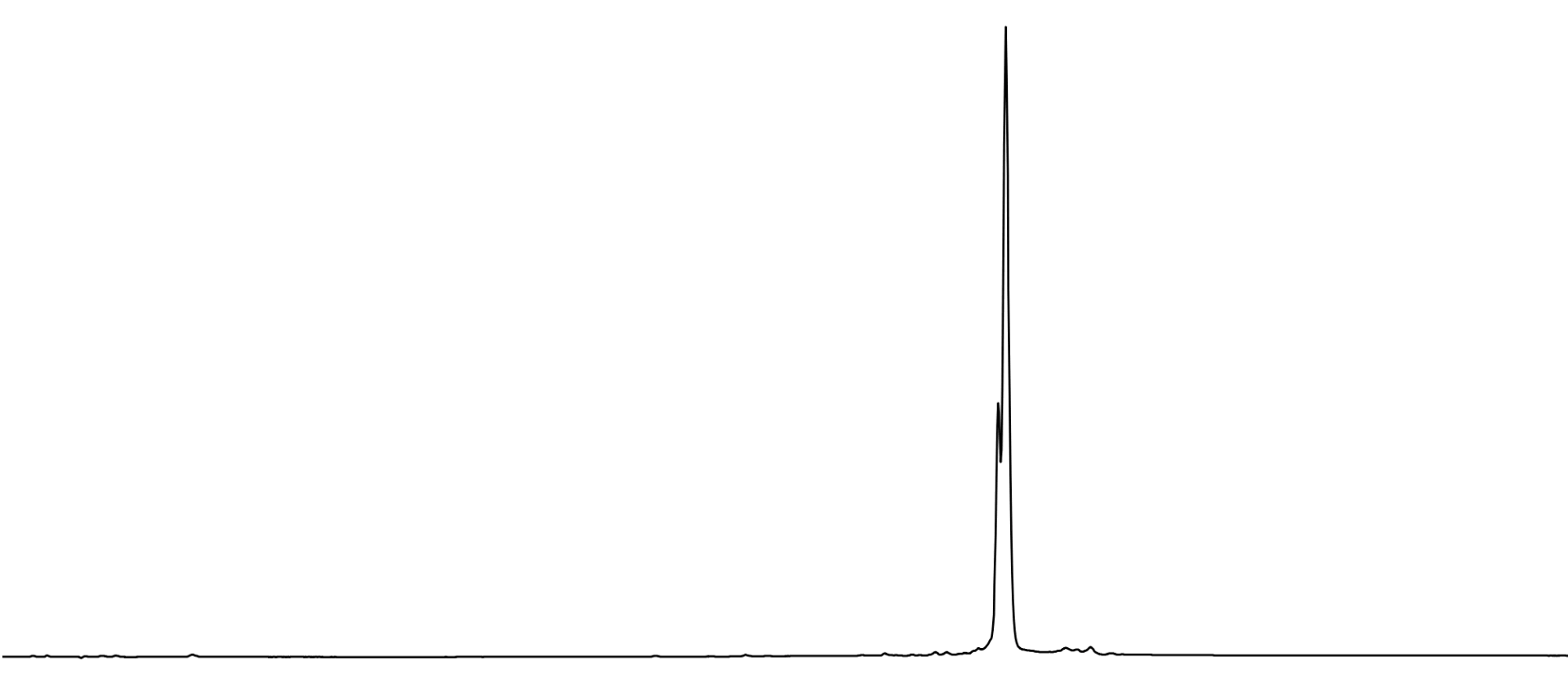
After the two step deprotection, the oligonucleotide was GlenPak™ purified by standard methods and analyzed by RP HPLC using a Waters X-Bridge column. The resulting chromatogram (Figure 4, Page 10) shows partially resolved diastereomers and a very clean oligo.
Figure 4. 5’-TXT TXT TXT TTT-3’, where X is 2’-OMe-U-PACE, deprotected as described and analyzed by RP HPLC using a Waters X-Bridge RP HPLC column.
However, when the oligo was analyzed by ESI MS, the deconvolved spectrum showed multiple peaks that corresponded to decarboxylation events and the loss of CO2 (-44 Da) totaling approximately 20% relative to the target mass (Figure 5A, Page 11). This is despite the fact there was no indication of the presence of methyl phosphonate impurities in the HPLC chromatogram. (Note, the small +303 Da peak is a trivial impurity due to incomplete removal of the 5'-DMT during GlenPak™ purification).
Upon review of the mass spectrum prior to deconvolution, an interesting observation was made - only the highly charged ions (-5 or higher) showed the presence of peaks corresponding to decarboxylation, suggesting these peaks were an artifact of the ESI MS. To test this hypothesis, the same sample was re-run using a lower voltage and the deconvolved spectrum showed an amazing improvement. The peaks corresponding to decarboxylation dropped to a mere 3.5% with the largest impurity being a +53 Da cyanoethylation peak resulting from the dT CEP monomers incorporated in the oligo (Figure 5B, Page 11).
Note that this cyanoethylation can be avoided by modifying the procedure to remove the cyanoethyl protecting groups
by first treating the support for 2-3 minutes with 1.5% DBU in acetonitrile, expelling the solution to waste, and then completing the 60 minute elimination using a fresh solution of 1.5% DBU.
Figure 5A. ESI MS of 5’-TXT TXT TXT TTT-3’ from Figure 4.
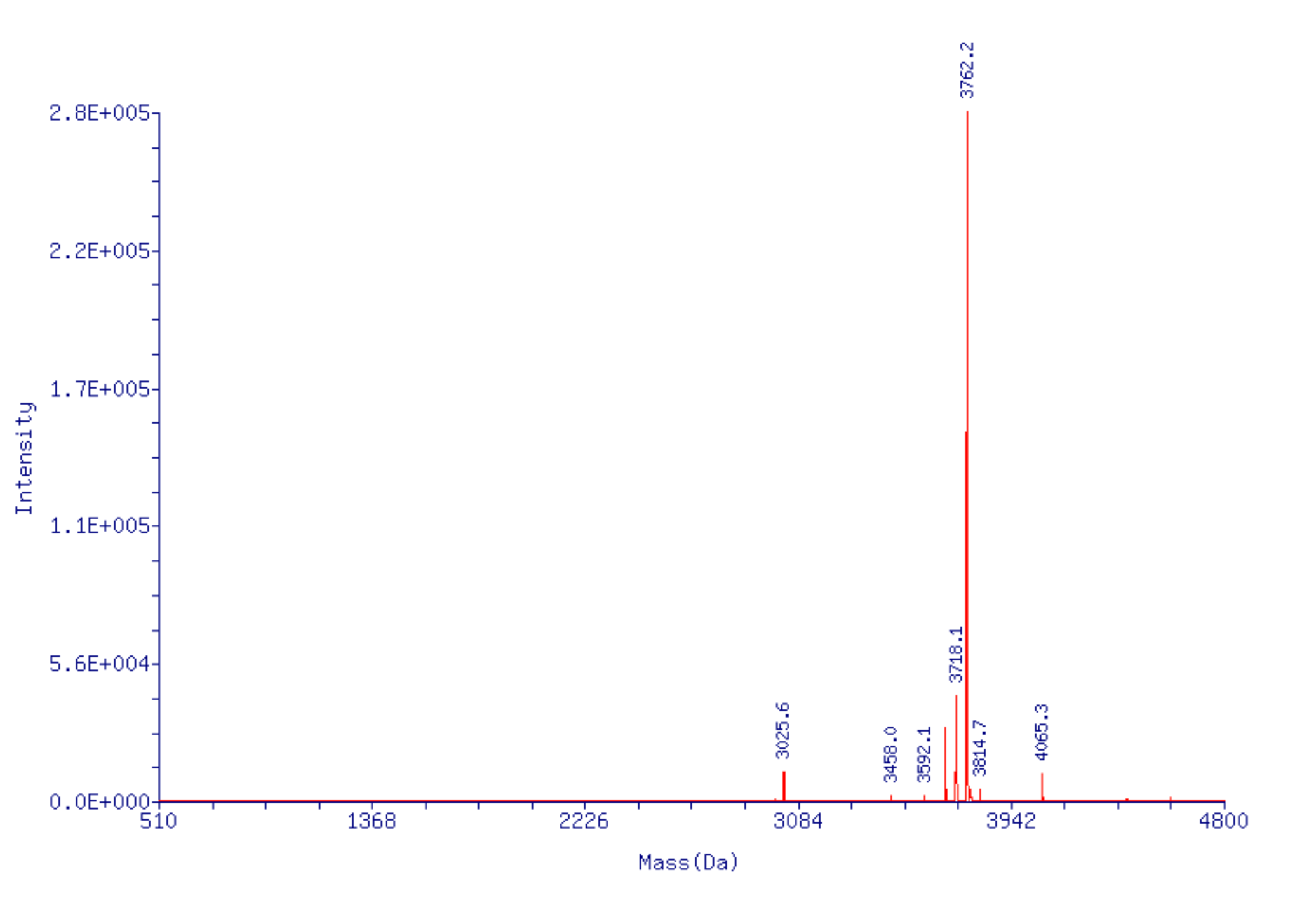
Figure 5B. ESI MS of 5’-TXT TXT TXT TTT-3’ repeated at lower voltage.
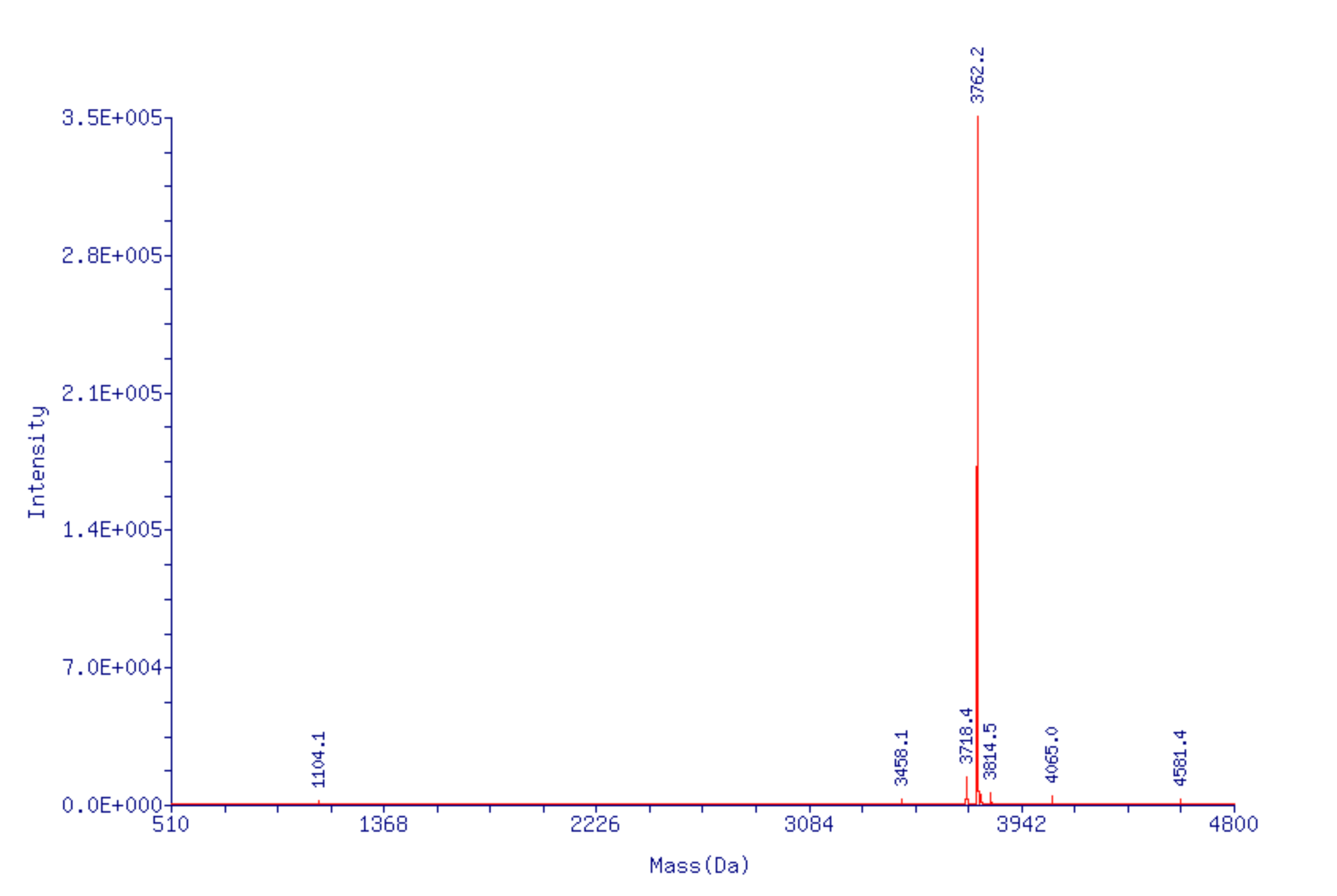
Figure 3. Structures of DNA and 2’-OMe-RNA PACE Monomers.
PLEASE SEE: Significant Improvement of CRISPR Specificity with 2'-OMe-PACE Modifications
Product Information
Pace products have been discontinued.
- Glen Report 30.11: Introducing ribo-tCo<>/sup>
- Glen Report 30.12: DBCO-Serinol Phosphoramidite
- Glen Report 30.13: Significant Improvement of CRISPR Specificity with 2'-OMe-PACE Modifications
- Glen Report 30.14: Use of 2'-OMe-PACE Monomers During Oligo Synthesis
- Glen Report 30.15: Methylene Blue II - A Unique Dye
- Glen Report 30.16: Technical Brief – 5'-Phosphorylation of RNA
- Glen Report 30.1: Technical Snippets