Glen Report 30.13: Significant Improvement of CRISPR Specificity with 2'-OMe-PACE Modifications
Authors: Zoltan Kupihara,b and Marvin H. Caruthersa
a Department of Chemistry and Biochemistry, Univ. of Colorado, Boulder, Colorado
b Department of Medicinal Chemistry, Univ. of Szeged, Hungary
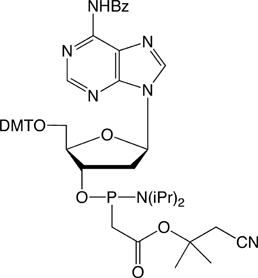
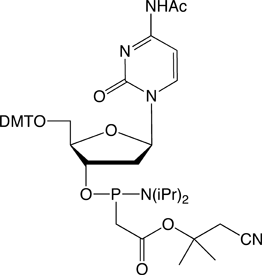
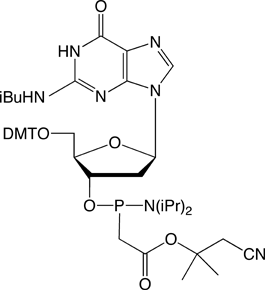
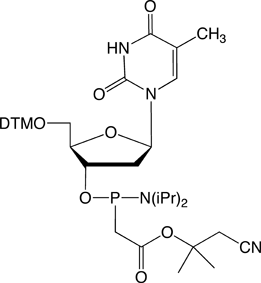
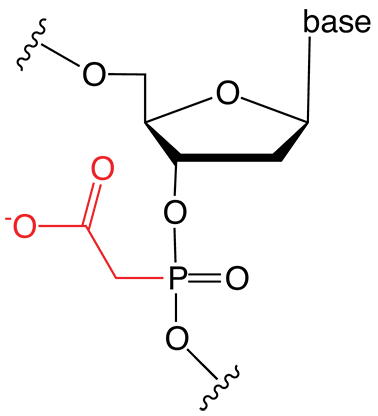
Figure 1. PACE Linkage
Introduction
Over the last 30 years there has been a vigorous search for modifications that enhance or enable nucleic acid therapeutics and diagnostics. The concept is that chemical modifications placed in specific positions can enhance the features of oligonucleotides and therefore impart therapeutic activity, while at the same time enhance the bioavailability through stabilization to nucleases and increased cellular penetration. The results of this search have essentially yielded two categories of chemical modifications showing wide utility:
- phosphorus modifications and
- modification at the 2'-position on the ribose sugar.
Modifications at the 2'-position can affect the binding characteristics to other nucleic acids and inhibit endonucleases but have limited effect on exonucleases, cellular penetration or bioavailability. Phosphorus modifications are useful to inhibit both endonucleases and exonucleases and have also been shown to increase bioavailability through association with serum proteins and enhanced cellular penetration.
Phosphonoacetate (PACE) Modification and Nuclease Resistance
One of the most recent phosphorus modifications to be developed is the phosphonoacetate (or thiophosphonoacetate) modification (PACE) which was reported first by Dellinger et al. in 2003.1 The PACE modification (Figure 1) contains a carbon-phosphorus bond and, like methylphosphonates, results in an internucleotide bond that is almost impervious to nucleases.
The ability of PACE modified oligonucleotides to impart nuclease resistance while retaining full or enhanced biological activity has been demonstrated.2 But unlike methylphosphonates, PACE backbone modifications retain the crucial nucleic acid recognition element of a negative charge on the backbone as a result of the carboxylate functional group. This negative charge, in addition to being an important recognition element for nucleic acid binding and function, makes the modification easy to incorporate into standard DNA or RNA chemical synthesis protocols and gives the resulting modified oligonucleotide typical solubility in aqueous solutions. PACE modified oligonucleotides allow efficient unassisted cellular uptake and significant miRNA inhibition effects without the need for transfecting agents.3 These interesting and easy modifications have been mostly overlooked based upon the timing of their discovery and a two-decade focus on increasing duplex binding affinity.
Effect of PACE Modification on Duplex Stability
In the initial publications on PACE modified oligonucleotides, the duplex stability experiments showed decreased Tm values for PACE modifications.4 In the case of DNA-DNA duplex experiments, the observed ΔTm change per linkage modification was -0.3 °C for the phosphonoacetate and -0.5 °C for the thiophosphonoacetate. When RNA was the complementary strand, the duplexes formed A-like DNA-RNA heteroduplexes also having decreased Tm values compared to natural DNA-RNA duplexes. The measured ΔTm per linkage modification were -1.3 and -1.8 °C for the phosphonoacetate and thiophosphonoacetate, respectively.
In 2012, the synthesis and biological evaluation of 2'-O-methyl-phosphonoacetate and thiophosphonoacetate RNA oligomers were also reported.3 According to the authors, the first two modifications incorporated, one at each end of the oligomer, increased the Tm by 0.9 °C for an 18-mer, but building in more modifications decreased the Tm by ~-1 °C per modification for phosphonoacetates and ~-0.75 °C per modification for thiophosphonoacetates. Later work5 using a 20mer RNA annealed to a DNA target strand gave a ΔTm of -1.0-1.1 °C per incorporation on average, whether the 2'-OMe PACE modifications were consecutively or non-consecutively inserted relative to unmodified RNA. As the thiophosphonoacetate, probes with consecutive incorporations were slightly more destabilizing with a ΔTm -1.3 °C compared to a ΔTm of -1.1 °C when the 2'-O-methyl thiophosphonoacetates were inserted non-consecutively within the sequence.
At the time the PACE modifications were introduced, there was a generally held belief that higher duplex stability could provide higher activity, and therefore PACE modifications were not considered as viable candidates for oligonucleotide therapeutics. That view began to change when researchers in the field of siRNA discovered that in certain circumstances decreasing duplex binding affinity decreased off-target activity, and increased on-target activity.6 As a result, a more modern view is that benefits can be obtained from modifications that either decrease or increase binding affinity. Such modifications can be used to enhance activity and increase specificity, thereby increasing the overall potency and decreasing the toxicity of oligonucleotide therapeutics. Most recently, it has become obvious that oligonucleotide therapeutics that are optimized to resist nucleases and target specific organs are having greater success in clinical trials.7
2'-OMe-PACE and CRISPR Gene Editing
Figure 2. 2’-OMe-PACE (inset) modified sgRNA lead to decreased off-target cutting of Cas9 in vitro.
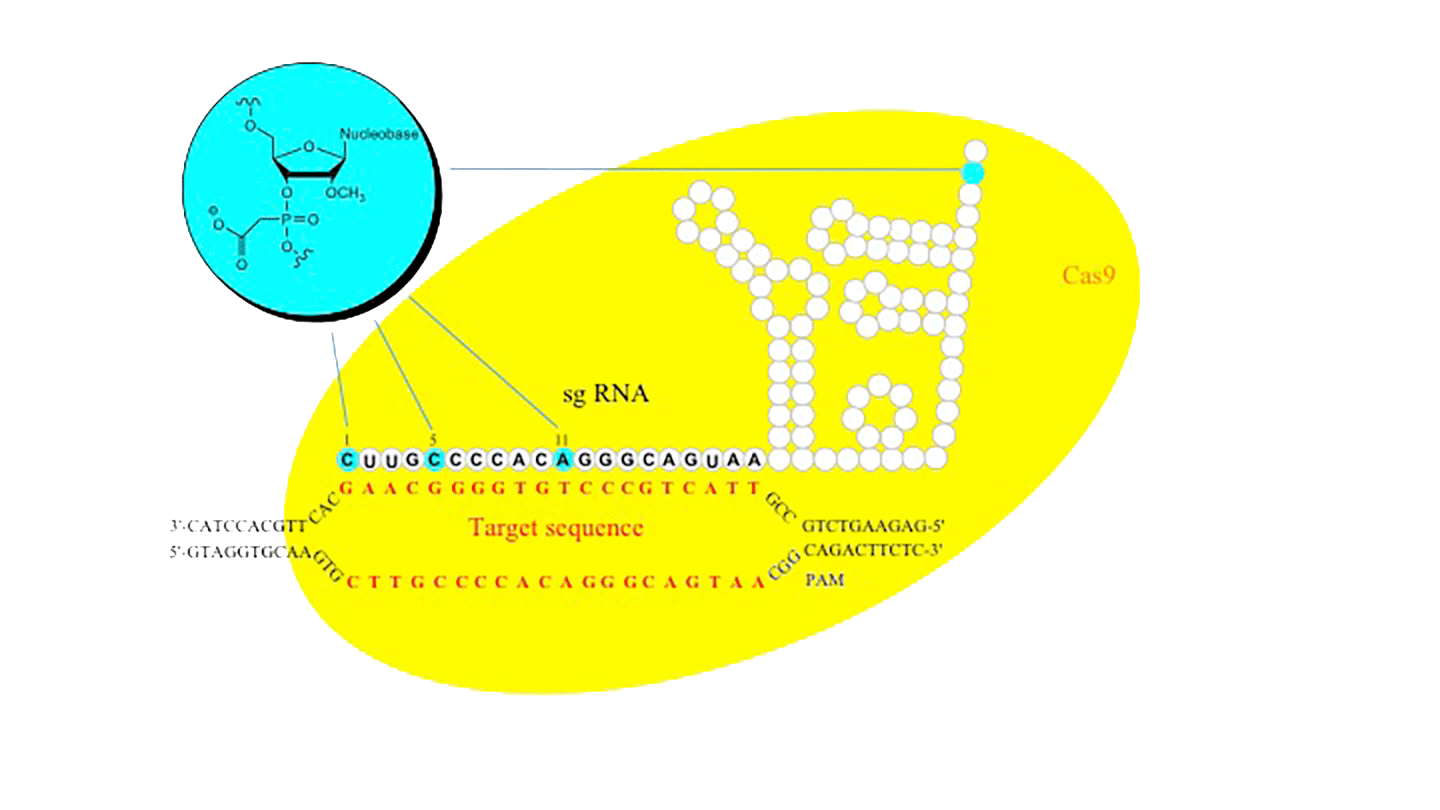
PACE modifications have enjoyed a resurgence in interest as applied to the field of CRISPR gene editing. In an initial publication, it was shown that single guide RNAs (sgRNA) provided significantly higher activity in cells when 2‘-O-methylthiophosphonoacetates were incorporated on the ends of the guide RNA to protect against cellular nucleases.8
In subsequent studies, 2'-OMe PACE modified single guide RNAs were also shown to significantly increase on-target specificity of the CRISPR-Cas9 (Streptococcus pyogenes) DNA cleavage in eukaryotic cells. In a recent paper, the incorporation of 2'-OMe PACE modified nucleotides in the 20-nucleotide guide region of the single guide RNA was shown (Figure 2) to decrease off-target cutting by over an order of magnitude while in most cases increasing the overall on-target efficiency as compared to unmodified single guide RNA.5
The authors utilized deep sequencing technology to evaluate off-target insertions and deletions (in/dels) at the known sites but also surveyed a library of 960 predicted potential off-target sites which had less than 6 mismatches to their initial intended target site. The authors observed a number of important results. The first was that, in the CRISPR-Cas9 system, the 2'-OMe PACE modification increased the sequence specificity and decreased the off-target cleavage. In addition, the 2'-OMe PACE modification also led to an overall increase of homology directed repair (HDR) at the desired site for gene editing. Finally, the authors found that the 2'-OMe PACE modification at positions 5 or 11 in the guide sequence gave the highest increase in sequence specificity across multiple gene targets, suggesting that this modification can be used ubiquitously in CRISPR systems to increase site-specific gene editing. The necessity for a high degree of sequence specificity in the field of Gene Editing is obvious; no one wants to create more mutations in the genome while trying to fix a specific mutation.9 However, it brings to light that increasing sequence specificity through chemical modifications is possible and will require a next generation of chemical modifications that can easily be incorporated into oligonucleotides using phosphoramidite chemistry.
Conclusion
Probably, the most remarkable characteristic of short oligonucleotides is their ability to differentially recognize and read the linear sequence of another oligonucleotide. No other biological molecule evolved this extraordinary ability. All hybridization based applications in modern biology utilizing chemically synthesized DNA and RNA rely upon this code selective binding phenomenon. This includes PCR, Sequencing, Probe Diagnostics, Array Hybridization, Antisense, siRNA, DNA barcoding, DNA storage, and many more.
We often take for granted that DNA and RNA have the unique ability to distinguish matched from mis-matched sequences, but there are limits. As you change the length, base composition and add modifications to an oligonucleotide, you change its ability to distinguish match from mis-match. In the early days of antisense therapeutics, there was a theory that “DNA Drugs” could be treated just like small molecule therapeutics and assumed they functioned as if the formation of a duplex to a target mRNA was analogous to a receptor/ligand binding event. That assumption led to the idea that tighter binding meant “better” activity. The result was a two decade-long search for “tighter binding” DNA and RNA modifications. Unfortunately, recognizing and preferentially binding to a target DNA or RNA sequence, and not binding to other similar sequences is a more complex event and may require less-tight binding modifications to achieve high target specificity, as shown in the recent CRISPR publication.5
As our knowledge of nucleic acid therapeutics has evolved it is clear that many early failures were the result of underestimating the need for a high level of nuclease resistance to allow for full therapeutic effect. As the field continues to evolve, highly nuclease resistant modifications, like PACE, may play a broader role in modification patterns used for therapeutics.
Glen Research is the exclusive provider of PACE modified monomers.
References
1. Dellinger, D. J., Sheehan, D. M., Christensen, N. K., Lindberg, J. G., and. Caruthers, M. H., J. Am. Chem.Soc., 2003, 125, 940–950.
2. Uhlmann, E. and M. Jurk, U.S. Patent No. 9,200,287 B2, 2015.
3. R.N. Threlfall, A.G. Torres, A. Krivenko, M.J. Gait, and M.H. Caruthers, Organic & Biomolecular Chemistry, 2012, 10, 746-754.
4. D. Sheehan, et al., Nucl Acid Res, 2003, 31, 4109-4118.
5. D.E. Ryan, et al., Nucleic Acids Res, 2018, 46, 792-803.
6. A. Khvorova, A. Reynolds, and S.D. Jayasena, Cell, 2003, 115, 209-16.
7. R.A. Haraszti, et al., Nucleic Acids Research, 2017, 45, 7581-7592.
8. A. Hendel, et al., Nat Biotechnol, 2015, 33, 985-989.
9. H. O'Geen, A.S. Yu, and D.J. Segal, Curr Opin Chem Biol, 2015, 29, 72-8.
PLEASE SEE: Use of 2'-OMe-PACE Monomers During Oligo Synthesis
Product Information
Pace products have been discontinued.
- Glen Report 30.11: Introducing ribo-tCo<>/sup>
- Glen Report 30.12: DBCO-Serinol Phosphoramidite
- Glen Report 30.13: Significant Improvement of CRISPR Specificity with 2'-OMe-PACE Modifications
- Glen Report 30.14: Use of 2'-OMe-PACE Monomers During Oligo Synthesis
- Glen Report 30.15: Methylene Blue II - A Unique Dye
- Glen Report 30.16: Technical Brief – 5'-Phosphorylation of RNA
- Glen Report 30.1: Technical Snippets