Glen Report 29.11: CDPI3 MGB™-Oligonucleotide Conjugates and Their Applications
Author: Eugene Lukhtanov
ELITechGroup Molecular Diagnostics
21720 23rd Drive SE, Suite 150
Bothell, WA 98021
Introduction
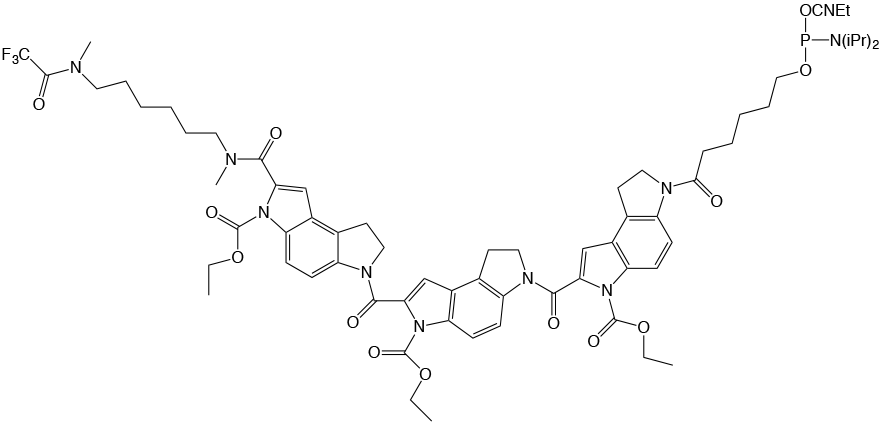
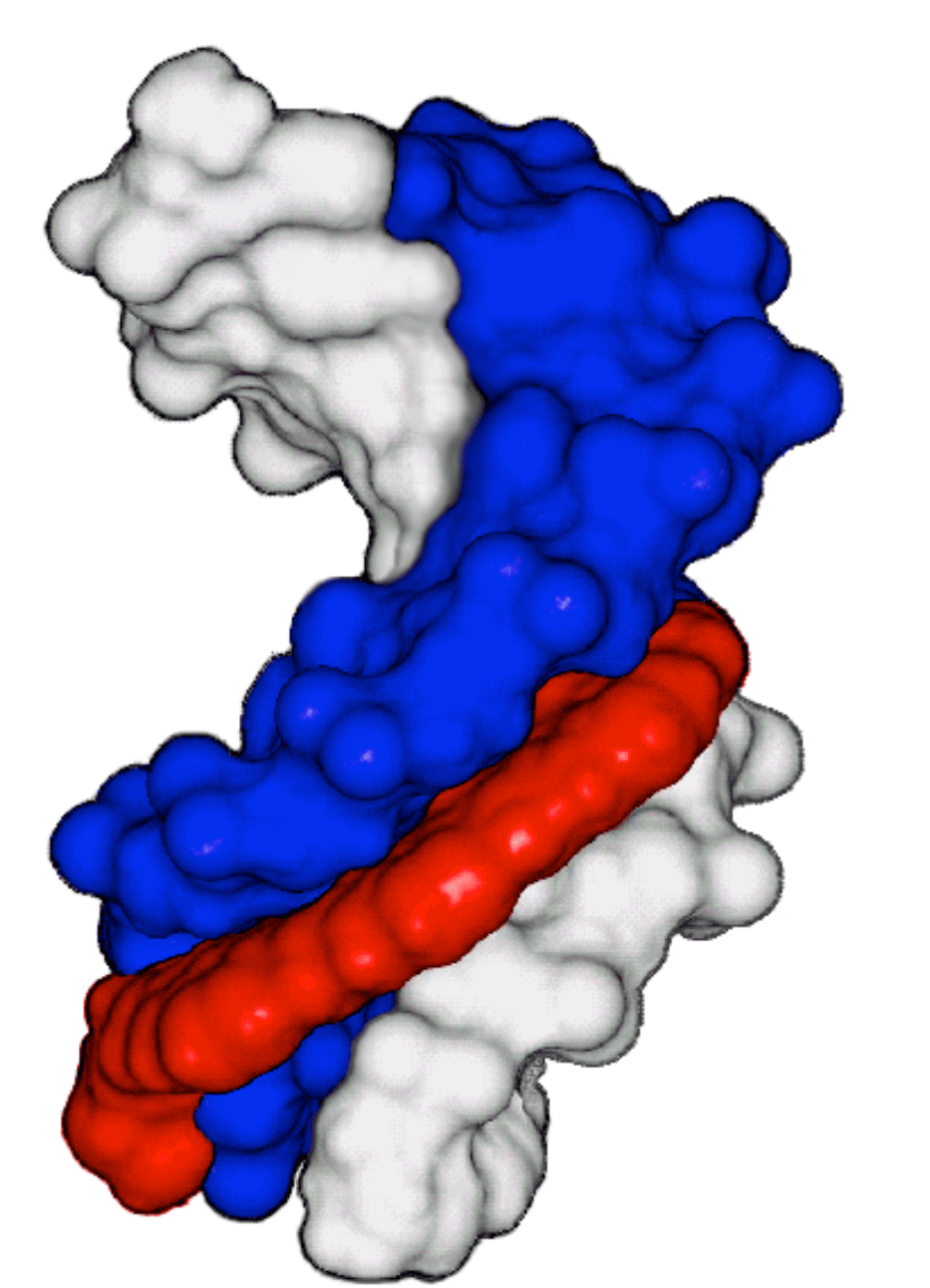
The tripeptide of dihydropyrroloindole-carboxylate (CDPI3)1 (Figure 1) is a minor groove binding (MGB) moiety derived from the natural product CC-1065 with strong DNA binding properties. CDPI3 is a crescent-shaped molecule which binds isohelically within the B-form DNA minor groove. The reversible binding is mediated via hydrophobic and van der Waals interactions between the MGB and the floor of the groove. CDPI3 moiety occupies a region of duplex DNA of approximately 5 bases long (Figure 2) and binds to both A/T and G/C rich sequences with association constants of Ka ~1x107 M-1 and Ka~1x105 M-1, respectively.
Figure 1: Structure of CDPI3 |
Figure 2: DNA duplex of CDPI3-ODN and Complement |
 |
 |
1,2-Dihydro-(3H)-pyrrolo[3,2-e]indole-7-carboxylate tripeptide (CDPI3) |
Connolly surface representation of a decamer DNA duplex formed between a CDPI3-ODN and its complement2. Red represents the CDPI3 and the linker moieties, Blue is the CDPI3-labeled strand and light gray is the complementary strand. |
Synthetic oligonucleotides (ODNs) with covalently-attached CDPI3 moieties were first introduced in 1995 by Lukhtanov et al.3 It has since been shown that such ODNs have enhanced DNA affinity and have improved the hybridization properties of sequence-specific DNA probes. Short CDPI3-oligonucleotides hybridize with single-stranded DNA to give more stable DNA duplexes than unmodified ODNs of similar length. For example, a DNA duplex formed between the CDPI3-(dTp)8 conjugate and poly(dA) template has a melting temperature (Tm) that is 44°C higher than the Tm of the unmodified duplex. Mismatch discrimination of short CDPI3-ODNs is also enhanced in comparison to unmodified oligonucleotides. It was demonstrated that mismatches under the CDPI3 binding region were more easily discriminated for a 15-mer CDPI3-ODN in comparison with unmodified ODN with up to 3-fold increase in free energy difference.4
The tethered CDPI3 moiety has a preference for A/T rich B-form DNA duplex but is capable of binding to DNA duplexes with mixed sequences as well as to some modified backbone nucleic acid as investigated by Kutyavin et al.5 The A/T sequence preference has a useful practical application. It is well known that DNA duplex melting temperature is dependent on A/T and G/C content with A/T rich sequences being much less stable. CDPI3 tethering significantly reduces the difference, such that probes of equal length have similar Tms regardless of base composition.
Applications
1. Arrest of primer extension and PCR blockers
CDPI3-ODN conjugates were investigated for potential use as antigene agents via the inhibition of DNA polymerase.6 The study, which was done in context of a single-stranded DNA phage, demonstrated that T7 DNA polymerase was physically blocked when a complementary 16-mer 5'-CDPI3-ODN was hybridized to a downstream site. Blockage was abolished when a single mismatch was introduced. A 16-mer with 3'-CDPI3 moiety also failed to arrest primer extension. The exceptional efficiency of the primer extension arrest was attributed to DNA polymerase's inability to displace the 5' end of the duplex super-stabilized with the CDPI3 group.
It has since been shown that 5'-CDPI3-ODNs are able to arrest Taq DNA polymerase and therefore can be used at PCR temperatures as well. This came as a surprise since it was expected that the 5' exonuclease Taq polymerase activity would degrade such duplexes. Evidently, 5'-CDPI3 labeling makes ODNs resistant to 5' exonuclease digestion. Such ODNs can be used as PCR blockers to prevent amplification of selected DNA sequences.
Figure 3: Mechanism of action of fluorogenic CDPI3-Fluorophore-primers |
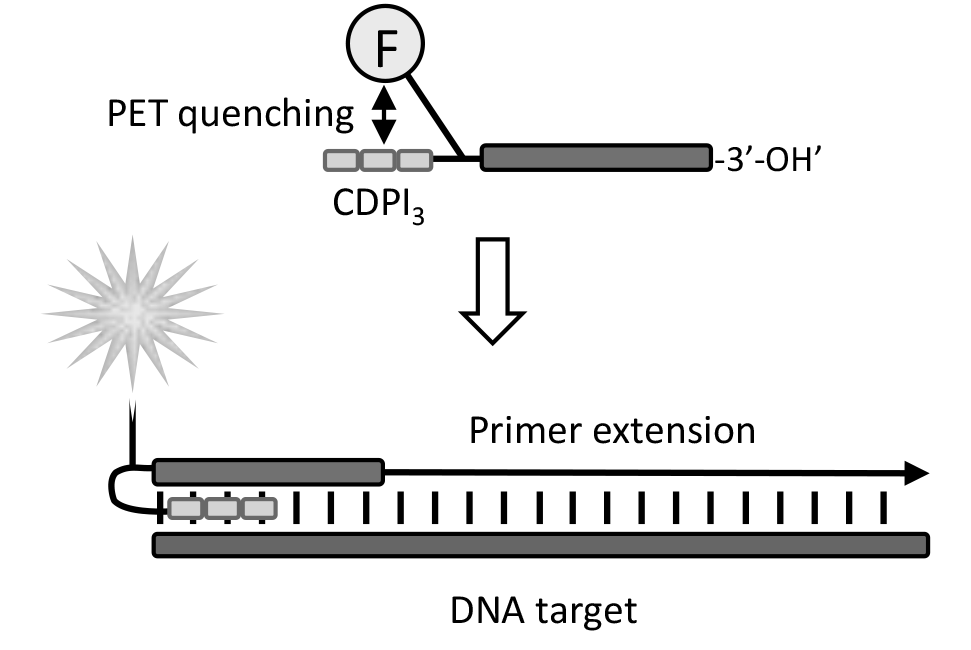 |
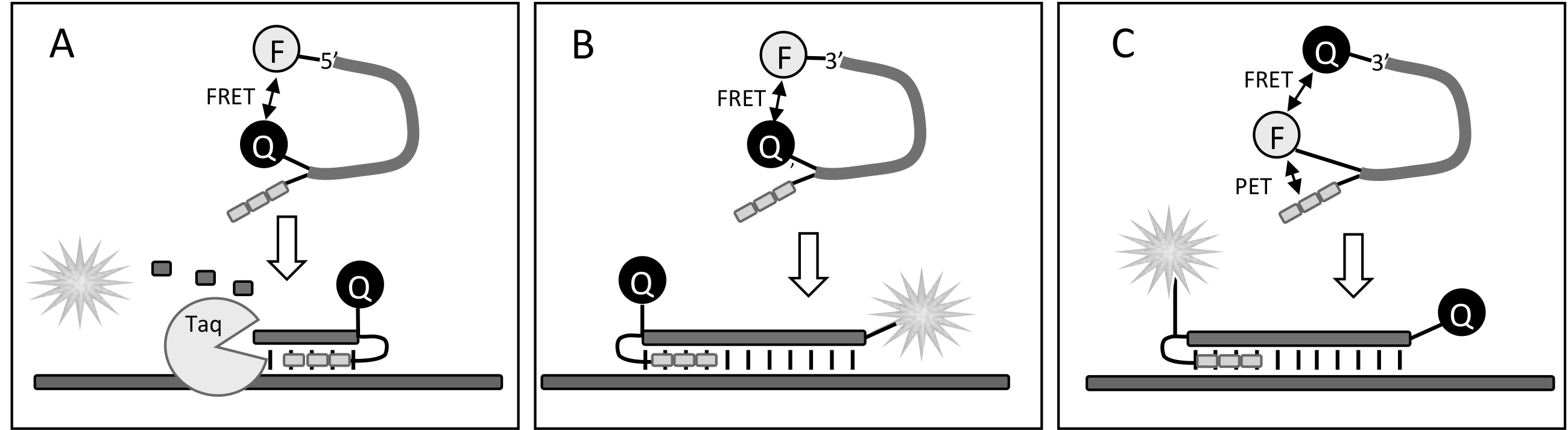
Figure 4: Putative mechanisms of action of MGB TaqMan (A), MGB Eclipse (B) and MGB Pleiades (C) probes |
 |
2. Short and fluorogenic PCR primers
Efficient priming of PCR was demonstrated with 5'-CDPI3-ODNs as short as 8-mers using modified (touch-down) PCR cycling conditions or 10-16-mers using regular PCR cycle.7 The PCR was shown to produce specific amplification products of expected size. The reduced length primers were suggested for use for PCR amplification of viral sequences which possess a high degree of variability or techniques such as gene hunting and differential display which amplify multiple sequences using short primer pairs.
5'-CDPI3-primers which also have an attached 5'-fluorophore are able to quench the dye fluorescence via the photo-induced electron transfer (PET) mechanism, depicted in Figure 3. Such primers are significantly quenched in a single strand state but become highly fluorescent when incorporated into the PCR amplicon allowing for detection of target amplification.
3. Real-time PCR probes
The stronger binding of CDPI3-ODNs in comparison with unmodified ODNs allows for more stringent hybridization conditions to be used in DNA hybridization assays. Short CDPI3-ODNs with improved mismatch discrimination are especially useful in PCR assays since they bind efficiently and specifically during the high-temperature primer extension cycle. Several types of real-time PCR probes that utilize the CDPI3 moiety have been developed.
- MGB TaqMan® probes4 have CDPI3-Quencher and Fluorophore tethered to the 3' and 5' ends, respectively. Provided that specific sequence is present in the target DNA, the TaqMan probes are degraded by Taq polymerase during PCR, releasing unquenched fluorophore.
- MGB Eclipse® hybridization probes8 have the CDPI3-Quencher and Fluorophore attached at the 5' and 3' ends, respectively. They are non-degradable and their fluorescence is strongly increased upon probes' hybridization to amplified targets during the annealing step.
- MGB Pleiades® probes9 are also non-degradable hybridization probes. They have the CDPI3-Fluorophore and Quencher moieties tethered to the 5' and 3' ends, respectively. These probes utilize the unique dual fluorescence quenching mechanism to significantly reduce background fluorescence and improve signal-to-background ratio.
4. miRNA Inhibitors
In a recent US patent,10 it was disclosed that CDPI3-ODNs with 2'-OMe sugar-phosphate nucleic acid backbone are highly efficient and specific miRNA inhibitors. The inhibition is more pronounced when the minor groove binder is tethered to the 5' end of an miRNA inhibitor. The mechanism of this effect is not fully understood but could be attributed to the abilities of 5'-CDPI3 moiety to stabilize nucleic acid duplexes and block enzymatic activities.
CDPI3-ODN-based miRNA inhibitors have also shown improved cellular uptake and promise as pharmaceuticals by modulating gene expression.11
References
[1] D.L. Boger, R.S. Coleman, and B.J. Invergo J.Org. Chem. 1987, 52, 1521-1530
[2] S. Kumar, M. Reed, H. Gamper, V. Gorn, E.A. Lukhtanov, M. Foti, J. West, R. Meyer Jr, B.I. Schweitzer, Nucleic Acids Res. 1998, 26, 831-838.
[3] E.A. Lukhtanov, I.V. Kutyavin, H.B. Gamper, and R.B. Meyer, Jr. Bioconjugate Chemistry,1995, 6, 418-426.
[4] I.V. Kutyavin, I.A. Afonina, A. Mills, V.V. Gorn, E.A. Lukhtanov, E.S. Belousov, M.J. Singer, D.K. Walburger, S.G. Lokhov, A.A. Gall, R. Dempcy, M.W. Reed, R.B. Meyer, J. Hedgpeth Nucleic Acids Res. 2000, 28, 655-661.
[5] I. V. Kutyavin, E. A. Lukhtanov, H. B. Gamper and R. B. Meyer, Jr. Nucleic Acids Res. 1997, 25, 3718-3723.
[6] I. Afonina, I. Kutyavin, E. Lukhtanov, R.B.Meyer, and H.Gamper. Proc. Natl. Acad. Sci USA, 1996, 93, 3199-3204.
[7] I. Afonina, M. Zhivarts, I. Kutyavin, E. Lukhtanov, H. B. Gamper, Jr., and R.B. Meyer, Jr. Nucleic Acids Res. 1997, 25, 2657-2660.
[8] I. Afonina, Y. Belousov, M. Metcalf, A.Mills, S. Sanders, I. Kutyavin, D. Walburger, V. Gorn, I. Shishkina, D. Adams, M. Ahmadian, R. Dempcy, D. Milesi, M.W. Reed, A. Vorobiev, A. Wald, N. Scarr, E. Yau, N. Vermeulen J. Clin. Ligand Assay, 2002, 25, 1-8.
[9] E.A. Lukhtanov, S. Lokhov, V.V. Gorn, M.P. Podyminogin, W. Mahoney Nucleic Acids Res. 2007; 35, No. 5 e30,doi:10.1093/nar/gkl1136.
[10] A. Khvorova, A. Vermeulen, R. Kaiser, J. Karpilow, N. M. J. Vermeulen, W. Mahoney US9334495 Minor groove binder (MGB)-oligonucleotide miRNA antagonists
[11] D. L. Mcelligott WO2012119051 Enhanced biodistribution of oligomers
Legal Notice
NOTICE TO PURCHASER: LIMITED LICENSE
This product is sold under licensing arrangements between ELITechGroup Inc. and Glen Research. The purchase price of this product includes limited, nontransferable rights to use the product solely for activities of the purchaser which are directly related to human diagnostics. Other uses, including incorporation of the product into another commercial product, are prohibited without additional license rights. For information on purchasing a license to this product for purposes other than those stated above, contact:
ELITech Group Molecular Diagnostics,
21720 23rd Drive SE, Suite 150,
Bothell, WA 98021.
Phone (425) 482-5555.
Fax (425) 482-5550.
Email: mdx@elitechgroup.com.
This limited license permits the person or legal entity to which this product has been provided to use the product, and the data generated by use of the product, only for human diagnostics. Neither ELITechGroup Inc. nor its licensors grants any other licenses, expressed or implied for any other purposes.
Some components of nucleic acid analysis, such as specific methods and compositions for manipulating or visualizing nucleic acids for analysis, may be covered by one or more patents owned by other parties. Similarly, nucleic acids containing specific nucleotide sequences may be patented. Making, using, offering for sale, or selling such components or nucleic acids may require one or more licenses. Nothing in this document should be construed as an authorization or implicit license to make, use or sell any so covered component or nucleic acid under any such patents.
Product Information
5'-CDPI3 MGB™ Phosphoramidite (10-5924)
- Glen Report 29.11: CDPI3 MGB™-Oligonucleotide Conjugates and Their Applications
- Glen Report 29.12: 5'-CDPI3 MGB™ Phosphoramidite and 3'-CDPI3 MGB™ CPG
- Glen Report 29.13: Studying DNA Repair Pathways Using Site-Specifically Modified Oligonucleotides
- Glen Report 29.14: N-Acetylgalactosamine (GalNAc) Oligonucleotide Conjugates
- Glen Report 29.15: Sequence Modification Using Glen Research's 5-Modified dU Family

