Glen Report 17.2: A Comparative Study of Commercially Available Universal Supports For Oligonucleotide Synthesis
A.V. Azhayev1, M.L. Antopolsky1, T.M.L.TennilÄ1, H. Mackie2, and J. B. Randolph21
University of Kuopio, Kuopio, Finland and 2 Glen Research Corporation, Sterling, VA, USA
A detailed comparison of seven commercially available universal supports, which have been examined for their ability to support the synthesis of short and long oligodeoxynucleotides (DNA oligomers) and also oligoribonucleotides (RNA oligomers), is reported herein. Our results demonstrate that the universal supports fall into two categories differentiated by the mechanism of elimination of the 3’-phosphate linkage to produce the desired 3’-hydroxyl group. In the first group, the oligomer is quickly cleaved from the support followed by slow dephosphorylation under aggressively basic conditions to generate the 3’-hydroxyl group. In the second group, the dephosphorylation step first leads to the cleavage of the oligomer from the support, followed by further deprotection of the oligomer under standard conditions. The first group is exemplified by McLean's1 classic support (1) and four of the seven supports tested fall into this category. The second group is more novel and the support that exhibited the most desirable features fell into this category. Support (5) was tested with CPG (5a) and polystyrene (5b) as the core particles. Universal support (5) proved to be the only truly universal support in that it was used successfully for the production of short and long DNA oligomers, as well as for the production of biologically active siRNA.
Introduction
Standard oligonucleotide synthesis uses a solid support that contains the first nucleoside covalently bound to the support by a linker that is hydrolyzed during the cleavage step following solid-phase synthesis. This support-bound nucleoside becomes the 3’-terminal residue of the final oligonucleotide after the cleavage and deprotection steps. Clearly, this approach requires the use of at least four solid supports for general DNA synthesis along with an additional four supports for RNA synthesis. Various solid supports containing unusual nucleosides for specific applications are also required.
A universal support does not have the intended 3’-nucleoside attached. Rather, the 3’-nucleoside or residue is added in the first cycle, generating an undesired phosphate linkage between this nucleoside and the universal support. This approach requires that this phosphate linkage be removed during the cleavage and/or deprotection steps. However, the universal support strategy offers the following clear advantages:
- eliminates the possibility of errors in parallel synthesis applications where up to 384 wells may contain different supports;
- eliminates the need for at least four supports for DNA synthesis and four supports for RNA synthesis;
- simplifies the preparation of oligonucleotides with modified or unusual nucleosides at the 3’-terminus.
Universal Supports
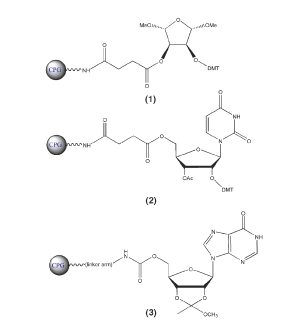
Several universal supports have been described in the literature1-5 and a selection of these is now commercially available. The structures of a selection of the commercially available supports are shown in Figures 1 and 2. These supports fall into two categories, as follows, depending on the timing of the dephosphorylation step that generates the 3’-hydroxyl of the target oligonucleotide.
Cleavage THEN Deprotection and Dephosphorylation
In the first category, the regular cleavage step of oligonucleotide synthesis predominantly leaves intact the residue or tether attached to the 3’-nucleoside through a phosphodiester linkage.
The dephosphorylation step along with elimination of the unwanted tether takes place during the deprotection step and usually requires stronger conditions than normal deprotection. These universal supports1 are either non-nucleosidic but incorporating a 5-membered ring similar to the ribose ring found in nucleosides or nucleoside-based supports, or protected nucleosides configured for base-mediated elimination. Examples of these supports are shown in Figure 1.
Cleavage by Dephosphorylation THEN Deprotection
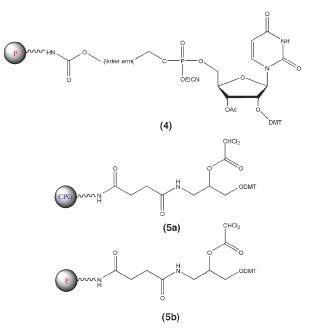
In the second group, the dephosphorylation step is the cleavage step and the only oligonucleotides released into solution already have a 3’-hydroxyl group. Further conventional deprotection leads to the fully deprotected oligonucleotide. The first example4 of this type of support uses a nucleotide attached to the support by a non-cleavable linker. The second example5 is a novel non-nucleosidic support. Examples of these supports are shown in Figure 2.
RNA Synthesis
Recent developments in RNA research, including the burgeoning use of siRNA, have led to an explosion in growth of oligoribonucleotide production. Instead of being carried out in a few specialist labs, RNA synthesis has now grown to a level requiring high-throughput synthesis and demands the use of a universal support. The first category of universal support defined above is incompatible with RNA deprotection since it relies on procedures known to degrade RNA. However, universal supports in the second category may be compatible with RNA synthesis.
Discussion
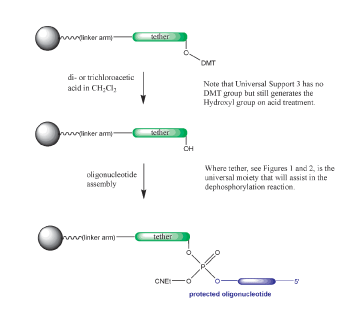
A number of universal supports have been introduced recently and representative structures are shown in Figures 1 and 2. All of these solid supports function similarly: regular detritylation (although universal support 3 has no DMT group, the regular deblock step generates the hydroxyl group), the addition of the first nucleoside monomer, and then the remaining oligonucleotide preparation steps proceed without any changes from standard procedures, as shown in Scheme 1.
Cleavage THEN Deprotection and Dephosphorylation
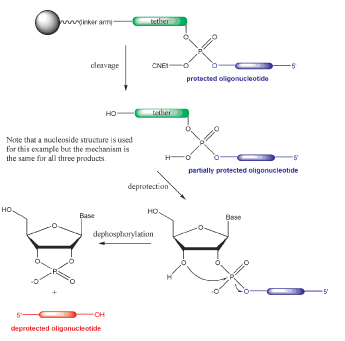
In the case of the supports in Figure 1, the elimination of the terminal phosphodiester group utilizes the same reagents (ammonium hydroxide, aqueous methylamine, a mixture of the first two (AMA), aqueous sodium hydroxide, etc.), as needed for routine deprotection of oligonucleotides, as shown in Scheme 2. However, much more aggressive and lengthy conditions are typically required. Upon the completion of oligonucleotide assembly, the 3’-terminal phosphotriester group is first converted into the phosphodiester function by ß-elimination of the cyanoethyl protection group, as shown in Scheme 2. Only upon the release of the 3’-hydroxyl of the tether nucleoside and hydrolysis of the linker to the CPG does the intramolecular nucleophilic attack on the phosphorous atom of the phosphodiester group take place to effect dephosphorylation. This dephosphorylation reaction is a relatively slow process, requiring lengthy aggressive treatment with ammonium hydroxide if the presence of some 3’-tethered product along with the target oligonucleotide in the final mixture is to be avoided. Thus, in the case of these supports, the process of oligonucleotide cleavage, base deprotection, 3’-dephosphorylation at elevated temperature and evaporation of aqueous ammonia normally require 8-10 h.
Another universal support 6 was also tested in this study but its structure was not revealed by the manufacturer. However, its behavior during testing indicated that it fell into this first category. Indeed, using the same tests as outlined in Table 1, this support also generated mixtures of the target oligomer and the 3’-tethered oligomer in ratios similar to those found for universal supports 1-3.
The product profile can be improved by adding metal ions to the mix, and Li+, Na+ and Zn2+ have all been used to speed up the elimination reaction, presumably by stabilizing the 5-membered transition state. However, the speed and simplicity of evaporation of the deprotection solution to give the crude oligonucleotide with no need for desalting is not possible with these ionic additives. All of these facts make these solid matrices unattractive for high throughput oligonucleotide manufacturing. These ionic additives would also assist in the degradation of RNA linkages and this makes their use to accelerate the dephosphorylation reaction especially unacceptable for RNA or siRNA production.
Cleavage by Dephosphorylation THEN Deprotection
When rationalizing the drawbacks of the first class of supports described above, two intrinsic problems must be emphasized. Firstly, the universal support should be designed in such a way that the process of cleavage/3’-dephosphorylation should release only the desired product. Secondly, the 3’-dephosphorylation reaction should proceed quickly. In other words, the processes of cleavage and 3’-dephosphorylation have to be, in essence, the same extremely fast process.
A nucleotide-based universal support (Support 4 in Figure 2) with a non-cleavable attachment to a polystyrene support offers a significant improvement over the supports described above. In this case, the universal linker is attached to the polymer via a phosphotriester group. Upon aqueous ammonium hydroxide treatment, this phosphotriester group, along with the oligonucleotide 3’-terminal phosphotriester group, are first converted into the phosphodiester functions by ß-elimination of the cyanoethyl protection groups, as shown in Scheme 3. Subsequent deprotection reactions are standard. Mechanistically, the intramolecular nucleophilic attack on the phosphorous atom of the 3’-phosphodiester group appears to be the rate-limiting step for the release of the target oligonucleotide into solution. The other phosphodiester group linking the universal tether to the support is stable under conditions of cleavage/dephosphorylation. As a result, even after heating at 60 °C for 8 h, only about 0.5% of 3’-tethered oligonucleotide is present in the mixture, along with 74% of 3’-dephosphorylated target oligomer, as shown in Table 2. This universal support affords reasonable quantities of a target oligomer, free from the 3’-tethered product, in a reasonable time and looks more attractive for high throughput applications than the first set described above.

The most recently described universal support (Supports 5a and 5b in Figure 2) may be the most likely to meet all of the criteria outlined above. The cleavage and 3’-dephosphorylation appear to be the same fast process (20 – 30 min), facilitated by a solution of anhydrous ammonia in methanol, as shown in Scheme 4. The labile dichloroacetyl group is cleaved prior to the ß-elimination of the cyanoethyl protection group of the phosphate moiety (closest to the spacer, linked to the solid matrix). This is followed by the rapid intramolecular nucleophilic attack on the phosphotriester function by the hydroxyl group. This reaction is additionally assisted by the neighboring amide function. All of these factors result in the very fast cleavage/dephosphorylation of the oligonucleotide, which is still predominantly nucleic base-protected. After removing the solid support, further deprotection procedures employ either standard protocols, e.g., addition of aqueous ammonium hydroxide to the solution of oligonucleotide in methanolic ammonia and heating the mixture at 55 °C for 5 h, or simply continued deprotection with methanolic ammonia (60 °C for 8 h).

.
It is noteworthy that a number of additional benefits were found using complete deprotection in anhydrous ammonia in methanol. Firstly, final evaporation of ammonia in methanol takes much less time than the evaporation of aqueous ammonia (4-5 times faster). Secondly, longer oligonucleotides (> 50mer) precipitate from the methanolic ammonia nearly quantitatively in the course of deprotection of nucleic bases. This allows separation of the product oligomers by centrifugation in less than 5 min.
Comparison of Universal Solid Supports for Oligonucleotide Synthesis
| Universal Support | Conditions of cleavage/deprotection (c/d) | Percentage of full length oligo with 3'-OH after c/d | Percentage of full length oligo with 3'-tether after c/d | Relative yield of all UV260 absorbing material after c/d# |
|---|---|---|---|---|
| 1 | 1 ml conc. NH3/H2O containing 15 mg of LiCl for 30 min at r.t., then 5h at 55°C | 45% | 31% | 82% |
| 2 | 1 ml conc. NH3/H2O containing 15 mg of LiCl for 30 min at r.t., then 5h at 55°C | 64% | 15% | 95% |
| 3 | 1 ml conc. NH3/H2O containing 15 mg of LiCl for 30 min at r.t., then 5h at 55°C | 42% | 35% | 70% |
| 4 | 1 ml conc. NH3/H2O for 30 min at r.t., then 5h at 55°C | 71% | 0.5% | 46% |
| 5a | 100 µl of 2M NH3/MeOH for 30 min at r.t., then 1 ml conc. NH33/H2O for 5h at 55°C | 83% | 0% | 98% |
| 5b | 100 µl of 2M NH3/MeOH for 30 min at r.t., then 1 ml conc. NH3/H2O for 5h at 55°C | 87% | 0% | 100% |
| 6* | 1 ml conc. NH3/H2O for 30 min at r.t., then 5h at 55°C | 58% | 22% | 82% |
| The oligonucleotide prepared was 5’-TTTTTTCACCGCCCGGTACACCCTTTTT-3’. # The yield of the target oligonucleotide, generated from the polystyrene Universal Support 5b, was taken as 100%. Contents of oligonucleotides in the crude mixtures were determined by ion-exchange HPLC. * The structure of Universal Support 6 was not disclosed by the manufacturer. |
||||
| Universal Support | Conditions of cleavage/deprotection (c/d)& | Percentage of full length oligo with 3'-OH after c/d | Percentage of full length oligo with 3'-tether after c/d | Relative yield of all UV260 absorbing material after c/d# |
|---|---|---|---|---|
| 1 | 1 ml of AMA 17h at 55°C | 66% | 0% | 85% |
| 2 | 1 ml of conc. NH3/H2O containing 15 mg of LiCl for 6h at 65°C | 79% | 0.3% | 71% |
| 3 | 1 ml of conc. NH3/H2O containing 17 mg of LiCl for 6h at 75°C | 77% | 0.5% | 93% |
| 4 | 1 ml of conc. NH3/H2O for 8h at 60°C | 74% | 0.5% | 74% |
| 5a | 1 ml of 3M NH3/MeOH for 8h at 60°C | 87% | 0% | 91% |
| 5b | 1 ml of 3M NH3/MeOH for 8h at 60°C | 87% | 0% | 100% |
| 6* | 1 ml of conc. NH3/H2O for 8h at 60°C | 81% | 1.3% | 73% |
|
The oligonucleotide prepared was 5’-TTTTTTCACCGCCCGGTACACCCTTTTT-3’. & Cleavage/deprotection conditions given in Table 2 were recommended by the manufacturers. # The yield of target oligonucleotide, generated from the polystyrene Universal Support 5b, was taken as 100%. Contents of oligonucleotides in the crude mixtures were determined by ion-exchange HPLC. * The structure of Universal Support 6 was not disclosed by the manufacturer. |
||||
Results
Materials and Methods
an ABI 392 DNA/RNA synthesizer employing its standard DNA synthetic protocol (1 µmolar scale) was used with the commercially available universal supports. Different protocols of cleavage/deprotection were used to generate oligonucleotides from these supports. Finally, the resulting reaction mixture was dissolved in water (1ml) and analyzed by ion-exchange HPLC on a DNAPacTM PA-100 (4x250) column using a linear gradient from 5 to 30% B in A for 30 min. (A- 0.1M sodium acetate in 20% acetonitrile; B- 0.1M sodium acetate and 0.4M sodium perchlorate in 20% acetonitrile).
DNA Synthesis on Universal Supports
In the first set of experiments, the supports were cleaved and deprotected using conditions constrained to 55 °C for heating for no more than 5 hours. These conditions are typically less aggressive than those recommended by the manufacturers but they gave an insight into the mechanism of dephosphorylation. Ion-exchange HPLC analysis of the crude mixtures reveals that the product oligonucleotide was usually present in two forms, the first fully deprotected and the second still containing the 3’-tether awaiting further dephosphorylation. The percentage of tethered product ranged from 15% to 35% of the crude oligonucleotide product. Analysis of the crude products from universal supports 4 and 5 reveal that virtually no tethered oligonucleotide is present in the mixture, demonstrating a different mechanism of dephosphorylation. The yields of all UV260 absorbing material were also determined from the ion-exchange HPLC data, These yields are recorded relative to the highest yield, obtained using universal support 5b. One other commercial support 6, whose structure was not revealed by the manufacturer, was also tested and the percentage of tethered product fell in the same range as supports 1-3, indicating that it is dephosphorylated using a similar mechanism. The results are summarized in Table 1.
In the second set of experiments, the universal supports were cleaved and deprotected following the manufacturers’ protocols. All supports performed well with the product oligonucleotide present in the crude mixture at levels ranging from 66% to 87%, with amounts of 3’-tethered product all falling below 1.5%. Again the yields of crude product were recorded relative to the universal support 5b, which generated the highest yield.
Finally, overall yields of various DNA oligomers (ranging in length from 20mer to 75mer) obtained from nucleoside bound CPG and Universal Supports 5a,b were basically the same. However, the yields of oligomers, prepared on supports 1-4,6, were always somewhat lower than the amount derived from nucleoside bound CPG and/or universal supports 5a,b.
RNA Synthesis on Universal Supports
Universal supports by definition should be appropriate for ALL types of oligonucleotide synthesis. In general, they make a lot of sense for DNA synthesis, but what is the state of play for RNA synthesis? From the results outlined in Tables 1 and 2, it is clear that the type exemplified by universal supports 1-3 is inappropriate for RNA synthesis. The dephosphorylation conditions are simply too aggressive for RNA. However, supports 4, 5 and 6 may be compatible with RNA synthesis.
For this study, two strands of siRNA were prepared as shown in Table 3. The RNA monomers were protected with the TOM protecting group6 and the conditions used for RNA deprotection had already been validated to produce biologically active siRNA. All four supports, 4,5a,b,6, gave RNA of purity ranging from 62% to 82%, as determined by ion-exchange HPLC. The yield from support 4 was low until more aggressive conditions were used for the cleavage/dephosphorylation step, which led to the lowest percentage of the target oligo in the crude mixture. Supports 5a and 5b gave good yields of crude RNA with reasonable purity of the target oligomer.
Comparison of Universal Supports in RNA# Synthesis
0.4ml of 4M NH3/MeOH 30 min at r.t., then 1 ml of 7M NH3/MeOH, 5h at 65°C, followed by TOM deprotection** 82% 100%<5a0.4 ml of 4M NH3/MeOH 30 min at r.t., then 1 ml of 2M Me-NH2/MeOH, 30 min at 65°C, followed by TOM deprotection** 79% 88%6*0.4 ml 32% NH3/H2O for 5h at 65°C, followed by TOM deprotection** 66% 54%6*0.4 ml of 32% NH3/H2O 30 min at r.t., then 1 ml of 40% Me-NH2/H2O, 30 min at 65°C, followed by TOM deprotection** 51% + 34% of 3'-tethered oligomer 66%5b0.4 ml of 4M NH3/MeOH 30 min at r.t., then 1 ml of 7M NH3/MeOH, 5h at 65°C, followed by TOM deprotection** 76% 100%5b0.4 ml of 4M NH3/MeOH 30 min at r.t., then 1 ml of 2M Me-NH2/MeOH, 30 min at 65°C, followed by TOM deprotection** 75%84%0.4 ml of 32% NH3/H2O 30 min at r.t., then 1 ml of 40% Me-NH2/H2O, 30 min at 65°C, followed by TOM deprotection** 82% 19%
| Universal Support | Conditions of cleavage/deprotection (c/d) | Percentage of full length oligo with 3'-OH after c/d | Relative yield of all UV260 absorbing material after c/d## |
|---|---|---|---|
| 5a | |||
| 4 | 0.4 ml 32% NH3/H2O for 5 h at 65°C, followed by TOM deprotection** | 62% | 61% |
| 4 |
Two strands of siRNA were synthesized in this study. The sense strand 5’-AGUCGCCUCGAAGAUACACtt-3’ was synthesized on CPG-based support 5a and 6, and the antisense strand 5’-GUGUAUCUUCGAGGCGACUtt-3’ was synthesized on polymeric supports 4 and 5b. Uppercase letters are given for the Ribonucleoside units and the lowercase t for the thymidine unit.
##The yield of target oligonucleotide, generated from the CPG Universal Support 5a with NH3/MeOH, was taken as 100% for oligos 1-4. The yield of target oligonucleotide, generated from the polystyrene Universal Support 5b with NH3/MeOH, was taken as 100% for the oligos 5-8. Contents of oligonucleotides in the crude mixtures were determined by ion-exchange HPLC.
* The structure of Universal Support 6 was not disclosed by the manufacturer.
** 0.5 ml of DMSO + 0.16 ml of HF/TEA, 65 °C, 30 min.; cool on ice and quench with 1 ml 0.1M Na acetate (sterile!!!), pH 5.2, 65 °C, 30 min.; cooled on ice, desalted and analyzed.
Use of Universal Support in the Synthesis of long DNA oligomers
| 75mer | Yield, AU260 |
|---|---|
| Seq 1* | 238 |
| Seq 2* | 171 |
| Seq 3** | 142 |
| Seq 4* | 238 |
| Seq 5* | 200 |
| Seq 6** | 118 |
Seq 1) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG AGA AGA AGA AGA AGA -3’
Seq 2) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG GAG GAG GAG GAG GAG -3’
Seq 3) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG AAG AAG AAG AAG AAG -3’
Seq 4) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG GAA GAA GAA GAA GAA -3’
Seq 5) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG GGA GGA GGA GGA GGA -3’
Seq 6) 5’-GAC CTG CAG GAA AAA AAA AAA GTA TGA GAG AGA GAT ATG TAT GTT TGG TAT TGG TTG TTG AGG AGG AGG AGG AGG -3’
Seq 2 was synthesized on 1000 Å dG-CPG and Seq 1,3,4-6 on 1000 Å Universal Support 5a.
*RP HPLC was used for oligonucleotide purification.
** Ion-exchange HPLC, followed by RP HPLC were used for oligonucleotide purification.
Synthesis of Longer DNA Oligomers on Universal Supports
While universal supports 5a and 5b performed best for DNA and RNA synthesis, is it reasonable to expect a universal support to be compatible with the synthesis of longer DNA oligos? An experiment was set up to compare the yield and purity of oligos prepared on a 1000Å CPG version of Support 5a with the synthesis on a conventional 1000Å deoxynucleoside support.
The results of this experiment are collected in Table 4. The product oligonucleotides Seq 1, 2, 4, 5 were purified DMT-on by reverse phase HPLC. The other two oligos, Seq 3, 6, were first purified DMT-off by ion-exchange HPLC followed by reverse phase HPLC. Yields were determined by measuring AU260.
As detailed in Table 4, it is clear that the universal support 5a is compatible with the synthesis of long oligos and good quality products can be obtained in good yield.
Conclusion
The main impediment to the universal adoption of a universal support has been the aggressively basic conditions required to complete the elimination reaction to release the terminal hydroxyl group. The standard reagents used in oligonucleotide deprotection are ammonium hydroxide and aqueous methylamine, which are popular since they are completely volatile. Using these reagents to carry out the elimination reaction requires either high temperature, with attendant high pressure, or extended reaction times. In addition, lithium chloride has been used to speed up the elimination reaction. However, the addition of salts to the deprotection solution requires an additional desalting step for the crude oligonucleotides and may be damaging to siRNA oligos.
The group of universal supports tested performed very well when used according to the manufacturers’ guidelines. However, the results outlined in Tables 1-4 show that only universal supports 4 and 5a,b are candidates to be truly universal. Universal supports 5a,b performed the best of the group, generating the best yields of oligonucleotide under the mildest conditions. This support type would be appropriate for the production of DNA oligos, long and short, as well as those requiring mild deprotection. It is also compatible with the synthesis of RNA and siRNA. The reagent used for the cleavage/dephosphorylation step is commercially available and the procedures described are fully compatible with high-throughput synthesis. Please contact Glen Research for further information.
References
- S. Scott, P. Hardy, R.C. Sheppard, and M.J. McLean, Innovations and Perspectives in Solid Phase Synthesis, 3rd International Symposium, 1994, 115-124.
- C. ScheuerLarsen, C. Rosenbohm, T.J.D. Jorgensen, and J. Wengel, Nucleosides and Nucleotides, 1997, 16, 67-80.
- A.V. Azhayev, Tetrahedron, 1999, 55, 787-800.
- M.H. Lyttle, D.J. Dick, D. Hudson, and R.M. Cook, Nucleosides and Nucleotides, 1999, 18, 1809-1824.
- A.V. Azhayev and M.L. Antopolsky, Tetrahedron, 2001, 57, 4977-4986.
- S. Pitsch, P.A. Weiss, L. Jenny, A. Stutz, and X.L. Wu, Helv Chim Acta, 2001, 84, 3773-3795.

