Glen Report 12.15: More Novel Monomers: Nucleoside α-Thiotriphosphates, 5-Hydroxy-Methyl-dU, Dabcyl Products, and a Highly Fluorescent Nucleoside
α-Thiotriphosphates
We continue to support the exciting novel technique of Nucleotide Analog Interference Mapping (NAIM). How else can you achieve RNA structural analysis with the resolution of X-ray crystallography with the simplicity and speed of a sequencing gel? Check out a couple of recent reviews1,2 of this method and be amazed at the power of the technique. We currently maintain a full set of A analogs as a-thiotriphosphates, the "monomers" used in NAIM.
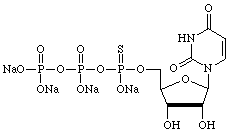
However, there are many other uses for α-thiotriphosphates so we have prepared, and will maintain, supplies of regular nucleoside and 2'-deoxy-nucleoside α-thiotriphosphates, as shown below.
| Adenosine | Cytidine |
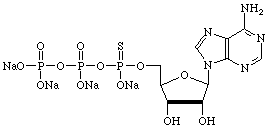 |
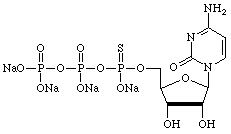 |
| Guanosine | Uridine |
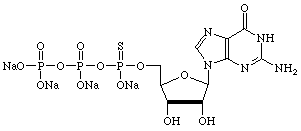 |
 |
| Inosine | 5-Methyl-Uridine |
 |
 |
| 2'-deoxyAdenosine | 2'-deoxyCytidine |
 |
 |
| 2'-deoxyGuanosine | Thymidine |
 |
 |
| 2'-deoxyInosine | |
 |
|
Product Information
The a-thiophosphates have been discontinued.
5-Hydroxymethyl-2'-deoxyUridine
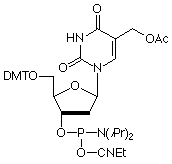
5-Hydroxymethyl-2'-deoxyUridine (HmdU) is formed from Thymidine by the action of peroxide radicals and ionizing radiation. This lesion may cause genetic mutations and chromosomal instability possibly by forming aberrant base pairs with G residues while maintaining its ability in a manner analogous to T to hydrogen bond with A. The cellular repair mechanism consists of excision of the mutant base by hydroxymethyluracil glycosylase to form apyrimidinic sites that are then repaired by excision repair. In our efforts to add to products to aid in studying DNA damage and repair, as well as mutagenesis, we are happy to add this monomer to our growing list of oxidatively damaged nucleosides.
Product Information
5-Hydroxymethyl-dU-CE Phosphoramidite (10-1093)
3'- and 5'-Dabcyl Group
The dabcyl moiety is fairly unique in its ability to quench the fluorescence of virtually any fluorescent tag in its immediate vicinity, making it the preferred quencher for molecular beacon probes. It should be noted that dabcyl quenching does not usually occur by a fluorescence resonance energy transfer (FRET) mechanism. Rather, the oligonucleotide probe is designed to be self-complementary so that the dabcyl group is in the vicinity of the fluorophore, providing quenching of signal until the probe finds its target. Only then does the probe become linear and, as the dabcyl group is separated from the fluorophore, quenching ceases and fluorescence is observed.
Typically, the dabcyl group is placed at the 3'-terminus and, for that setup, we offer 3'-dabcyl CPG 1000. This support will allow probes up to 60mer in length to be readily prepared. Since the dabcyl group is at least as hydrophobic as any fluorophore, we also offer 5'-dabcyl phosphoramidite. With the most hydrophobic group at the 5'-terminus, it is easier to purify the doubly-labelled oligonucleotide by reverse phase cartridge or HPLC. For example, a probe prepared with fluorescein at the 3'-terminus and dabcyl at the 5'-terminus is considerably easier to purify than the opposite setup.
5'-Dabcyl Phosphoramidite (10-5912)
3'-Dabcyl CPG (20-5912)
A Novel Fluorescent Nucleotide: Furano-dT
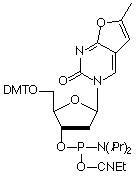
Fluorescence Resonance Energy Transfer (FRET) is a powerful tool for elucidating DNA structure and dynamics by probing both the distance and orientation between donor and acceptor fluorophores. However, this yields information regarding DNA structure only indirectly. An interesting alternative would be to excite the bases within the DNA strand itself and observe the energy transfer to a fluorophore. Our most recent fluorescent nucleotide analog, Furano-dT, allows one to do just that. Upon unwinding of a hybridized strand, a five-fold increase in fluorescence is observed at 470 nm when exciting at 260 nm. However, when exciting at 350 nm, no hybridization dependence is seen, which should allow the determination of relative population states. Also, because this fluorophore has only two additional carbon atoms, it may be well tolerated by DNA binding proteins.
References
- S.P. Ryder and S.A. Strobel, Methods, 1999, 18, 38-50.
- S.A. Strobel, Curr Opin Struct Biol, 1999, 9, 346-52.
Product Information
Furano-dT has been discontinued
- Glen Report 12.11: Oligonucleotide Dendrimers: From Poly-Labelled DNA Probes to Stable Nano-Structures
- Glen Report 12.12: TOM-Protected Minor Base RNA Phosphoramidites
- Glen Report 12.13: Poly-Pak Purification of Labelled Probes
- Glen Report 12.14: MerMade Instruments
- Glen Report 12.15: More Novel Monomers: Nucleoside α-Thiotriphosphates, 5-Hydroxy-Methyl-dU, Dabcyl Products, and a Highly Fluorescent Nucleoside
- Glen Report 12.16: Product Update - Which Chemical Phosphorylation Reagent?



