Glen Report 12.11: Oligonucleotide Dendrimers: From Poly-Labelled DNA Probes to Stable Nano-Structures
M.S. Shchepinov
Dept. Of Biochemistry
University of Oxford
Introduction
Dendrimers are discrete, highly branched, monodispersed polymers that possess patterns reminiscent of the branching of trees. Dendrimer oligonucleotides are representative of a new segment of polymer science1.
Two strategies for the synthesis of oligonucleotide dendrimers are possible: divergent - with the structure growing from the center to the periphery; and Convergent - with growth from the periphery to the center. The number of branches generated at each step determines the number of repetitive steps necessary to build up the desired molecule and the density of groups at the periphery. The groups or moieties on its outer shell determine the properties of the molecule.
Applications proposed for dendrimers exploit the high density and the large number of groups on the periphery. For example, dendrimers with a positively charged surface interact strongly with nucleic acids, helping to transport them through the membranes of cells2; dendrimers with internal cavities interact non-covalently with guest molecules to give guest-host systems3. In oligonucleotide chemistry, branched oligonucleotides4 were used for signal amplification in hybridization tests, making it possible to detect the target at a level of below 100 molecules/mL5.
Synthesis of Oligonucleotide Dendrimers
Described here is a set of branching phosphoramidite synthons which can expand the potential of oligonucleotide synthesis, opening ways to 2D and 3D structures. The set, shown in Scheme 1, consists of symmetric and asymmetric doublers (1) and (2)6 and a symmetric trebler (3)7. These phosphoramidite products permit the synthesis of a dendrimeric structure on top of a conventional "monomeric" oligonucleotide, as well as directly on the solid support. The monomeric and the dendrimeric sequence segments can be prepared with different lengths and different orientation by using 3'- and 5'- nucleoside phosphoramidites. The branches can terminate in any moiety available from the arsenal of phosphoramidite synthons. In this way using current synthesis technology, the symmetric doubler and trebler can be easily exploited to develop molecules like the symmetrical examples shown in Scheme 2, with a variety of desirable features.
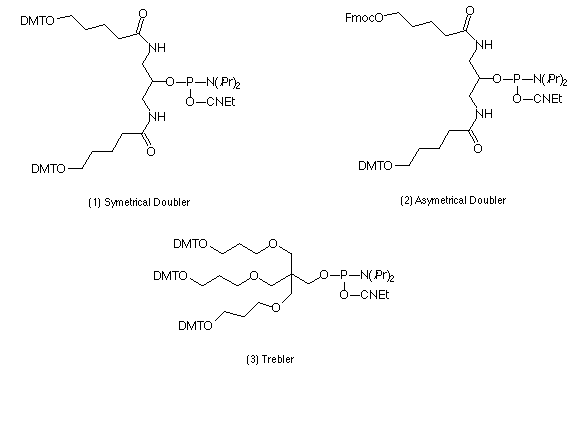
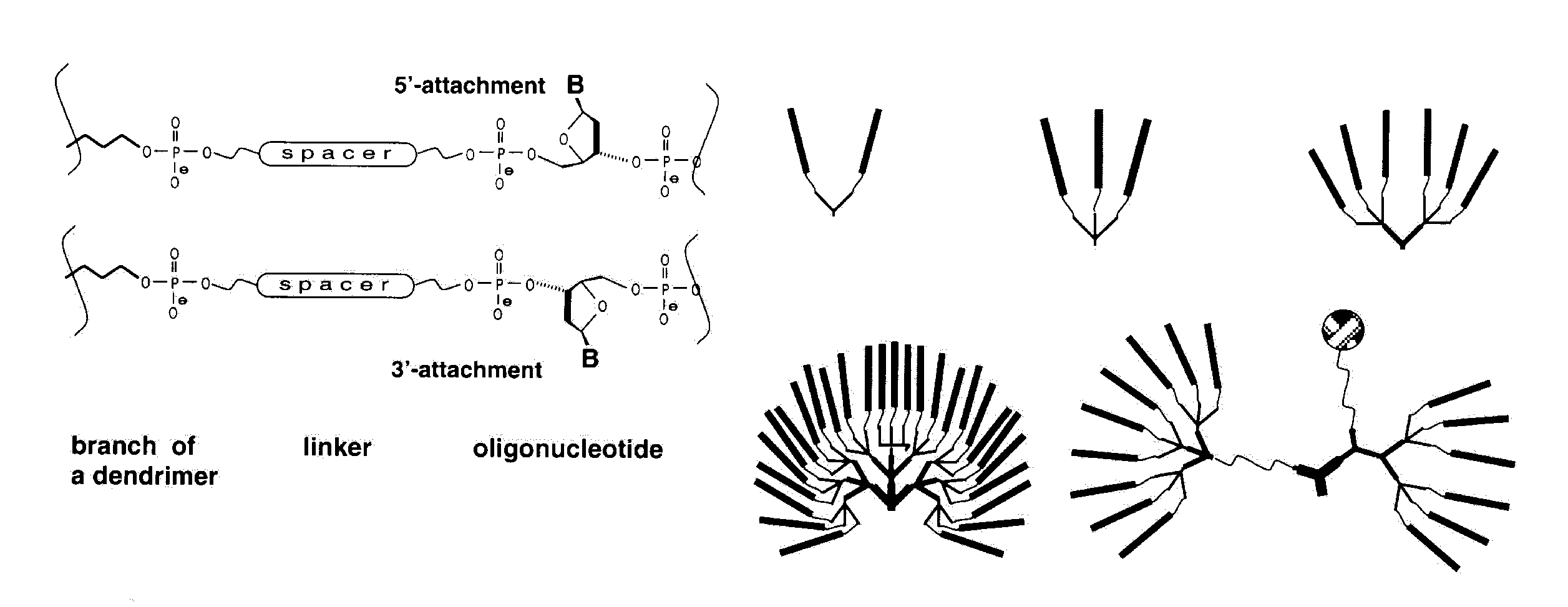
Previously, an asymmetric branching reagent6 has been used to prepare comb structures which offer good control of the number and spacing of hybridization sites. Novel applications for dendrimers could also be opened up if mixed terminal functionalities (e.g., oligonucleotides, reporter groups) were introduced onto the outer surface of dendrimers to give asymmetric structures like the example shown in the lower right of Scheme 2. These can be introduced at any generation of dendrimer synthesis using the asymmetric doubler (2), which contains two primary hydroxy groups protected with Fmoc and DMT. If different chemistries are then applied along the two different branches opened by different protecting groups, the result will be a dendrimer with two different functionalities on the surface and (possibly) different internal links. The generation at which the asymmetric doubler is introduced will determine the relative contribution of the two functionalities. After addition of the asymmetric doubler, its DMT-protected arm is first used to build up a desired structure. After final detritylation and capping the Fmoc group is deprotected8 and the synthesis is then carried out to generate the second half of the structure, with subsequent final deprotection.
When using 5-10 min condensation time, the doublers (1) and (2) give high condensation yields (>95%) for up to 4 condensations. The trebler (3) gives stable trebling for up to 3 condensations when using 500Å CPG supports, and 4-5 condensations using a 2500Å CPG support, thus giving ca 80-240 terminal hydroxyl groups. Elongation of branches using propan-1,3-diol or ethyleneglycol-based linkers further improves the yield of the next condensation with the trebler, as does adding the spacer between the first trebler and the solid support.
Spacer phosphoramidites added directly after the branching phosphoramidites can be used to reduce the density of packing of the dendrimer branches7. Spacers can also affect the size, flexibility, solvation properties and chemical functionality of the dendrimers.
Labelled Oligonucleotide Dendrimers
Dendritic oligonucleotides bearing several biotin residues, fluorophores or radioactive isotopes can be detected at much lower concentrations due to multiple labelling. Such amplification of signal may be particularly important in in situ hybridization and in the emerging techniques which exploit oligonucleotide arrays9, 10, where the signal is limited by the surface density of the device. But too dense a concentration of reporter groups leads to self-quenching of fluorescence. It also makes some oligonucleotide branches unavailable for hybridization and renders them inaccessible to bulky enzymes. In these circumstances the use of spacers is necessary.
Dendrimers containing 'bouquets' of short oligonucleotides can be labelled with g-32P-ATP and polynucleotide kinase. When used as probes to oligonucleotide arrays, the multiply-labelled structures show much higher sensitivity than their mono-labelled counterparts. The large dendrimeric structure does not affect the hybridization yield, even with oligonucleotides tethered to a surface9,10,11,12.
Besides the obvious means of amplifying the signal, it is possible, using synthons with different protective groups, to introduce different labels on the ends of different branches. These could be fluorophores with different emission spectra, providing the potential for a wide palette of colors; or the different ends could bear the donor and acceptor of an energy transfer pair; or just a different sequence on each branch.
Oligonucleotide Dendrimers as PCR Primers
A range of applications is opened up when dendrimeric oligonucleotides are incorporated into products by PCR to prepare, for example, probes with higher labelling capacity. Dendritic primers are indeed compatible with PCR conditions. The branched structure has a considerable effect on the electrophoretic mobility in that the PCR products have reduced mobility in gel electrophoresis. Products of PCR are double-stranded and therefore make poor hybridization probes without further treatment. Many of the methods for making probes, such as an asymmetric PCR, are cumbersome. The structure of the dendrimer is such that it leaves a 'bouquet' of oligonucleotides as a 5'-overhang on the double-stranded PCR product. Such a structure is naturally resistant to a number of exonucleases, and double-stranded PCR products are readily converted to single strands by T7 Gene 6 exonuclease7, as shown in Scheme 3.
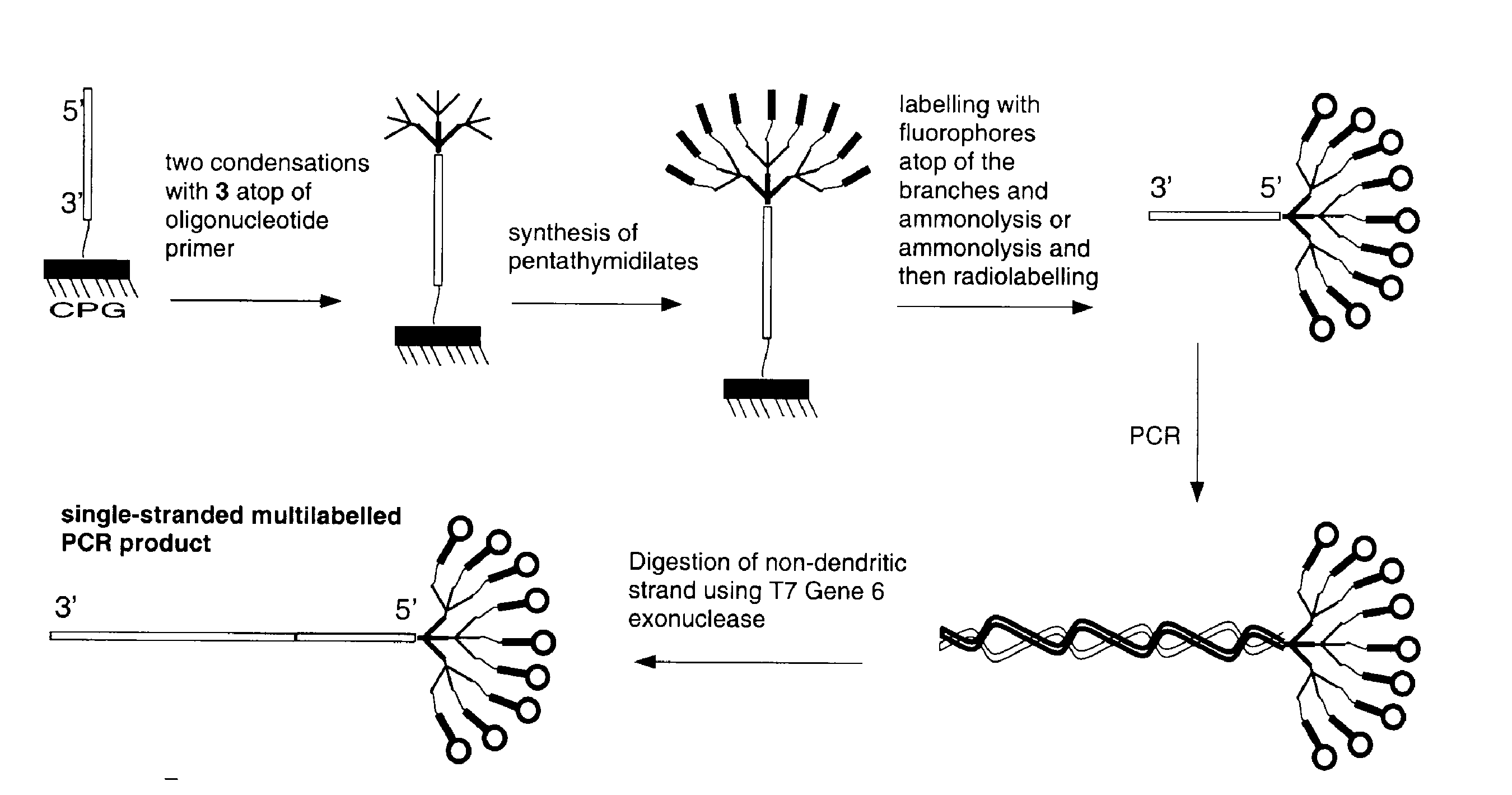
Mobility Shift by Dendrimeric Tags
It can be useful to modify the electrophoretic mobility of a PCR product by attaching a polymeric tag to one of the primers. The resulting mobility shift can be used to spread a set of multiplexed PCR products so that they do not interfere with each other. The addition of a dendrimeric 'bouquet' of short oligonucleotides to a PCR primer has a profound effect on the mobility of the PCR product. Addition of 9 pentathymidines decreased the mobility in agarose gels by an amount equivalent to adding 50 base pairs. This effect is even larger in polyacrylamide gels because of the smaller pore size. The number of couplings required to make the branched tag is small, especially when compared with the number of couplings required to make an unbranched polymer with the same effect on mobility. A total of 7 steps are used to make the dendrimeric tag: 50 steps would be needed to add 50 bases.
Nano-structures from DNA Dendrimers
Controllable formation of nano-scale architectures in solution and on solid supports is central to a range of activities in the emerging field of nanotechnology13. DNA molecules are well suited for these purposes because of their unique molecular recognition features. For example, dendrimers with arms terminating in oligonucleotides of the same or of different sequences could be used to build cages, cryptands, tubes, nets, scaffolds and other more complex 3-D structures11, shown in Scheme 4.
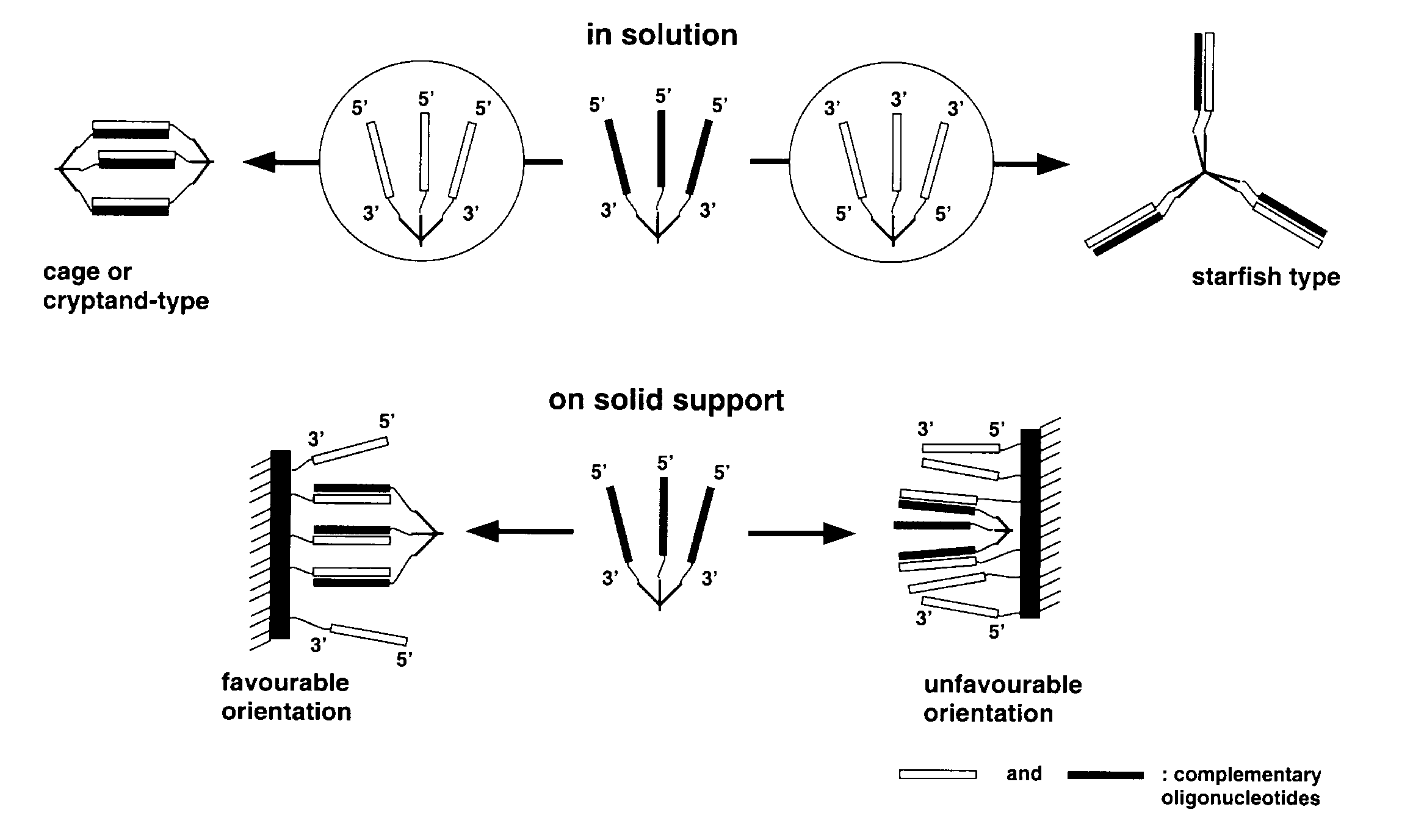
A different range of structures is possible when dendrimers interact with oligonucleotides bound to a solid support, which essentially represent a dendrimer with an infinite number of branches and a single core. In this case, the formation of an infinite network is not possible. When all oligonucleotides on the surface are involved in duplex formation, the complements will form a monolayer on the surface. However, growth of structures from the surface can be seeded from dendrimers which have some branches used to tie to the surface and others which are not complementary to the oligonucleotides on the surface. Complex patterns of oligonucleotides with different sequences can readily be synthesized on surfaces, for example, by light directed methods12 or a physical masking method9,10.
Duplexes formed from dendritic DNA have unexpectedly high thermal stability; the melting temperature is substantially higher than that of linear counterparts. It is an effect of combining the binding forces of individual duplexes by tying their ends together in the dendritic structure.
Conclusions
- Plain and mixed oligonucleotide dendrimers can be synthesized using novel doubling and trebling phosphoramidite synthons.
- Incorporation of label using g-32P-ATP and polynucleotide kinase increases in proportion to the number of 5'-ends.
- Fluorescent signal also increases in proportion to the number of 5'-ends, if spacers are incorporated between the labels and the ends of the branches.
- When using a dendrimeric oligonucleotide as a PCR primer, the strand bearing the dendrimer is resistant to degradation by T7 Gene 6 exonuclease making it easy to convert the double-stranded product of the PCR to a multiply labelled, single-stranded probe.
- For DNA dendrimers of different generations reassociated as complementary pairs in solution or with an array of complementary oligonucleotides on a solid support, duplex stability is greater than that of unbranched molecules of equal length. Enhanced stability of DNA dendrimers makes them useful as building blocks for the 'bottom up' approach to nano-assembly.
- These features also suggest applications in DNA chip technology when higher temperatures are required, for example, to melt secondary structure in the target.
References:
- G.R. Newcome, C.N. Moorefield, and F. Vogtle, Dendritic Molecules: Concepts, Synthesis, Perspectives; VCH Publishers: 1996.
- O. Boussif, et al., Proc Natl Acad Sci U S A, 1995, 92, 7297-301.
- J.F.G.A Jansen, et.al., Science, 1994, 266, 1226.
- T. Horn and M.S. Urdea, Nucleic Acids Res, 1989, 17, 6959-67.
- M.L. Collins, et al., Nucleic Acids Res, 1997, 25, 2979-84.
- M.S. Shchepinov and E.M. Southern, Russ. J. Bioorg. Chem., 1998, 24, 794.
- M.S. Shchepinov, I.A. Udalova, A.J. Bridgman, and E.M. Southern, Nucleic Acids Res, 1997, 25, 4447-4454.
- C. Lehmann, Y.Z. Xu, C. Christodoulou, Z.K. Tan, and M.J. Gait, Nucleic Acids Res., 1989, 17, 2379.
- E.M. Southern, et al., Nucleic Acids Research, 1994, 22, 1368-1373.
- E.M. Southern, U. Maskos, and J.K. Elder, Genomics, 1992, 13, 1008-17.
- M.S. Shchepinov, K.U. Mir, J.K. Elder, M.D. Frank-Kamenetskii, and E.M. Southern, Nucleic Acids Res, 1999, 27, 3035-41.
- A.C. Pease, D. Solas, E.J. Sullivan, M.T. Cronin, C.P. Holmes, and S.P. Fodor, Proc Natl Acad Sci U S A, 1994, 91, 5022-6.
- G.M. Whitesides, J.P. Mathias, and C.T. Seto, Science, 1991, 254, 1312-9.
Product Information
- Glen Report 12.11: Oligonucleotide Dendrimers: From Poly-Labelled DNA Probes to Stable Nano-Structures
- Glen Report 12.12: TOM-Protected Minor Base RNA Phosphoramidites
- Glen Report 12.13: Poly-Pak Purification of Labelled Probes
- Glen Report 12.14: MerMade Instruments
- Glen Report 12.15: More Novel Monomers: Nucleoside α-Thiotriphosphates, 5-Hydroxy-Methyl-dU, Dabcyl Products, and a Highly Fluorescent Nucleoside
- Glen Report 12.16: Product Update - Which Chemical Phosphorylation Reagent?

