Glen Report 11.24: Nucleotide Analogs for Interference Mapping of RNA
Scott Strobel
Dept. of Molecular Biophysics and Biochemistry, Yale University
Introduction
Single-site nucleotide analog substitution has been a commonly used approach for gaining atomic resolution biochemical information about RNA function. In this approach, a specific chemical group is modified or deleted at a defined position within an RNA prepared by solid phase chemical synthesis. The oligoribonucleotide is then assayed for its ability to perform the biochemical function of the native molecule. Although a powerful method, it is limited by the technical difficulty of synthesizing and characterizing a series of singly substituted RNAs. This is particularly true of longer RNA molecules (>40 nucleotides), which cannot be routinely synthesized as a single oligoribonucleotide. Here we describe an alternate approach to rapidly screen in vitro transcribed RNAs for biochemical effects resulting from functional group modifications.
Interference Mapping
Nucleotide Analog Interference Mapping (NAIM) is a chemogenetic approach that makes it possible to simultaneously, yet individually, probe the contribution of a particular functional group at almost every RNA nucleotide position in a single experiment1. The method utilizes a series of 5'-O-(1-thio)nucleoside analog triphosphates in a modification interference procedure that is as simple as RNA sequencing. In a NAIM experiment the smallest mutable unit is not the base pair, but rather the individual functional groups that comprise the nucleotides. Because the modification or deletion of a particular functional group within an RNA can severely affect its activity, this approach makes it possible to efficiently determine the chemical basis of RNA structure and function.
Instead of synthesizing a series of RNAs with chemical substitutions at specific sites, NAIM utilizes a combinatorial approach. Each nucleotide analog is prepared as a triphosphate for incorporation into the RNA during DNA templated in vitro transcription. The nucleotide analogs used in NAIM include a specific chemical alternation to the base or sugar, and an a-phosphorothioate substitution which serves as a chemical tag. The nucleotide analog triphosphate is randomly incorporated into an RNA transcript, where the phosphorothioate linkage can be selectively cleaved by the addition of I2 to produce a series of RNA cleavage products whose lengths correspond to the sites of analog incorporation2. By radioactively or fluorescently tagging one end of the RNA transcript, cleaving the RNA with I2, and resolving the cleavage products on a denaturing polyacrylamide gel, the sites of analog incorporation throughout the RNA can be individually assayed and used for interference analysis. The phosphorothioate tagged nucleotide analogs make it possible for all of the positions in the RNA to be assayed individually for functional group modification in a single experiment.
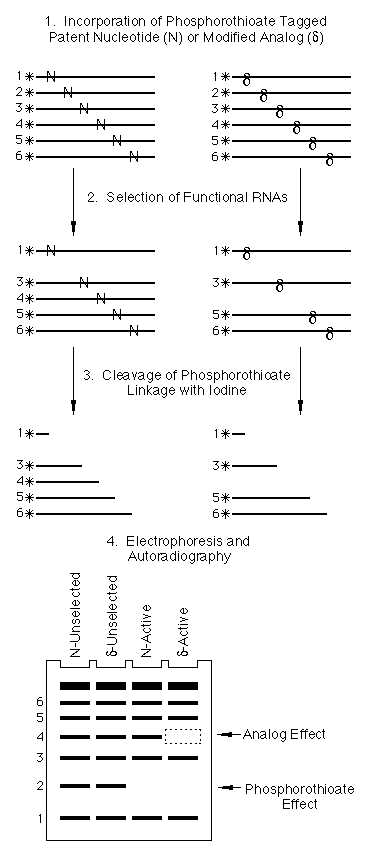
Because the phosphorothioate chemical tag is independent of the nucleotide analog whose location it reports, NAIM is generalizable to any analog that can be incorporated into a transcript by an RNA polymerase. A typical NAIM experiment is comprised of four steps ( Fig. 1) (i) The phosphorothioate tagged nucleotide analog is randomly incorporated throughout the RNA to create a family of transcripts, each of which contains only a few substitutions. A different transcription reaction is performed for each analog. (ii) The functional RNA variants in the population are separated from the inactive transcripts. The exact nature of the activity assay is specific for the RNA being studied, but could include affinity chromatography, native gel electrophoresis, filter binding, selective radiolabeling, etc. (iii) The phosphorothioate linkages in the active and unselected RNA populations are cleaved by I2 addition to mark the sites of analog incorporation within each molecule. (iv) The individual RNA fragments are resolved by gel electrophoresis and visualized by autoradiography. Sites of analog substitution that are detrimental to function are scored as gaps in the sequencing ladder among the active RNA variants (Fig. 1). Because every position in the sequence is a unique and independent band on the sequencing gel, a single screen can define the effect a particular analog has at every incorporated position within the RNA. The approach is applicable to any RNA that can be transcribed in vitro and has an assayable function that can be used to distinguish active and inactive variants. RNA functions that are amenable to this approach include catalysis, folding, protein or ligand binding, and the ability to act as a reaction substrate.
Nucleotide Analogs
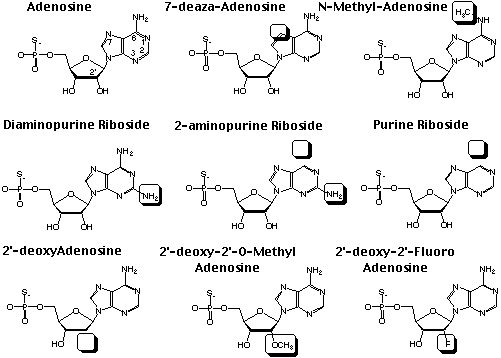
NAIM utilizes a -phosphorothioate tagged nucleotide analogs, each of which includes an incremental chemical alteration in the base or ribose sugar. The most completely developed set of analogs are those of adenosine, for which eight different analogs have been utilized in NAIM (Fig. 2)3. Five analogs modify the nucleotide base and three modify the ribose sugar. The base analogs include purine riboside (PuraS), N6-methyladenosine (m6AaS), tubercidin (7dAaS), diaminopurine riboside (DAPaS), and 2-aminopurine riboside (2APaS). The ribose sugar analogs all modify the 2'-OH group and include 2'-deoxyadenosine (dAaS), 2'-deoxy-2'-fluoroadenosine (FAaS), and 2'-O-methyladenosine (OMeAaS). All of the analogs can be randomly incorporated into an RNA transcript at an ideal 5% level of efficiency using either the wild-type T7 RNA polymerase or a Y639F RNA polymerase point mutant4. Each of these analogs provides specific information about the chemical basis of RNA activity at almost every incorporated position in the transcript. A similar collection of analogs can be utilized for the other three nucleotides G, C, and U.
PuraS, 2APaS and m6AaS measure the effect of modifications to the N6 exocyclic amine of adenosine. PuraS and 2APaS delete the amine, and m6AaS replaces one proton of the amine with a methyl group. m6AaS interference indicates that either both hydrogen atoms of the amine are necessary, or that there is insufficient space in the local structure to accommodate the additional methyl group. PuraS and 2APaS interference identifies sites where the amine is important for activity.
Interference with 7dAaS is indicative of an important major groove contact to the ring nitrogen, as this nucleotide replaces the N7 nitrogen with a CH group. Interference with PuraS, m6AaS, and 7dAaS are strong indicators of Hoogsteen hydrogen bonding.
DAPaS and 2APaS both add an additional amine to the C2 position of adenosine. In general, DAPaS and 2APaS show interference in areas of close packing in the minor groove of RNA. Another characteristic effect observed with DAPaS is enhancement of activity when paired with a U in regions of the molecule where duplex stability is important for function. OMeAaS also serves as a probe for tight packing in the minor groove.
dAaS interference identifies the 2'-OH groups important for RNA function, while FAaS delineates the role these 2'-OH groups play as either hydrogen bond donor or hydrogen bond acceptors. If a 2'-OH shows interference with both analogs, it suggests that the 2'-OH is a hydrogen bond donor. If instead the position has dAaS, but not FAaS interference, it argues that the 2'-OH at this site is a hydrogen bond acceptor.
In a few cases FAaS interference can also provide indirect information about the conformation of the ribose sugar for a given nucleotide. FAaS interference at sites lacking dAaS interference suggests that the 2'-OH does not make a direct contribution to activity. Instead, it argues that there is an indirect effect due to the chemical nature of the fluorine substitution. The 2'-fluoro group is highly electronegative, and as such, the ribose sugar favors the C3'-endo conformation5. This indirect effect is consistent with sites of C2'-endo ribose sugar conformation in a defined RNA system3, and may provide a biochemical signature for sites with unusual sugar pucker in other RNAs.
As our initial entry into this new collection of reagents, we are glad to offer a set of the adenosine analogs shown in Fig. 2. The collection will be expanded in the future to include a series of analogs for each of the nucleotides. We expect these molecules will become powerful reagents for the identification of the chemical groups important for a wide variety of RNA and DNA functions.
References
- S. A. Strobel and K. Shetty, Proc. Natl. Acad. Sci. U.S.A., 1997, 94, 2903-2908.
- G. Gish and F. Eckstein, Science, 1988, 240, 1520-1522.
- L. Ortoleva-Donnelly, A. A. Szewczak, R. R. Gutell and S. A. Strobel, RNA, 1998, 4, 498-519.
- R. Sousa and R. Padilla, EMBO J., 1995, 14, 4609-4621.
- S. Uesugi, H. Miki, M. Ikehara, H. Iwahashi and Y. Kyogoku, Tetrahed. Lett., 1979, 42, 4073-4076.
Product Information
The a-phosphorothioates have been discontinued.
- Glen Report 11.21: Advances in RNA Synthesis and Structural Analysis
- Glen Report 11.22: TOM-Protecting-Group™ - A Major Improvement in RNA Synthesis
- Glen Report 11.23: New Columns for Expedite and LV Applications
- Glen Report 11.24: Nucleotide Analogs for Interference Mapping of RNA
- Glen Report 11.25: Alternatives to Expedite Monomers
- Glen Report 11.26: More Novel Monomers