Epigenetics is the study of heritable changes in gene expression and regulation that are not due to changes in the DNA sequence itself. Rather, epigenetic control primarily involves methylation of cytidine in CpG islands and the modification of histones. Aberrant methylation of DNA is associated with a variety of disorders such as Beckwith-Wiedermann, Prader-Willi, and Angelman syndromes, as well as cancers such as neuroblastoma, Wilms tumour, and osteosarcoma, among many others. A few of the chronic diseases associated with dysregulation of DNA methylation patterns include cardiovascular disease, obesity, lupus, and certain types of leukemia.
Until recently, 5-methyl-2’-deoxyCytidine (5-Me-dC) was the only known modification of DNA for epigenetic regulation. In 2009, however, Kriaucionis1 discovered a second methylated cytidine, 5-hydroxymethyl-2’-deoxyCytidine (5-hmdC). While most prominent in neuronal tissues such as the brain and spine, 5hmdC has also been identified in a broad range of tissues such as the bladder, heart, liver and pituitary glands.2 At present, its role in the epigenetic control of gene expression remains unclear. However, since it prevents the targeting of a number of transcriptional repressors specific for 5-Me-dC, it appears to serve a subtle regulatory role.3 Removal of the methyl group from 5-Me-dC (demethylation) also plays an important role in cellular reprogramming, embryogenesis, autoimmune disorders and is critical in the establishment of maternal and paternal methylation patterns.
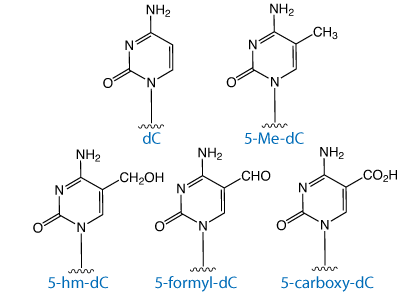
DNA demethylation occurs through active and passive mechanisms. Passive mechanisms simply involve the synthesis of DNA with the absence of methylation. Active DNA demethylation is thought to occur through a number of different mechanisms such as base excision repair (BER), nucleotide excision repair (NER), or the direct removal of the methyl group from 5-Me-dC.4 Direct demethylation from cytidine requires the breaking of a carbon-carbon bond through an unidentified mechanism. More likely, the removal of the methyl group involves the hydroxylation of 5-Me-dC to 5-hmdC or deamination of 5-Me-dC to thymidine and subsequent BER. An alternative demethylation pathway could involve the oxidation of 5-Me-dC to 5-formyl-dC or 5-carboxy-dC with subsequent decarboxylation to cytosine, though these active demethylation intermediates were not detected using a sensitive HPLC-MS assay.2,5
We are pleased to introduce two new cytidine analogs, 5-carboxy-2’-deoxyCytidine (dCCOO) (1) and 5-formyl-2’-deoxyCytidine (dCFo) (2) as CE phosphoramidites (Figure 1) that complement our existing tools for epigenetics research. We hope they will aid in the development and understanding of this exciting field.
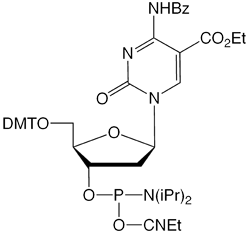

Despite the extra negative charge of the 5-carboxylic acid, the incorporation of 5-carboxy-dC into a DNA duplex has a stabilizing effect on DNA as indicated by a modest increase in the Tm (~2 °C per incorporation).6 The 5-carboxy moiety of cytidine projects into the major groove of a duplex, minimizing the potential negative steric effect of the modification. In comparison, the incorporation of 5-carboxy-dC into triplex forming oligos (TFO) as the third strand has a destabilizing effect, lowering the melting temperature by approximately 4-5° C. This is most likely because of the repulsion of the extra negative charge with the phosphate backbone and the crowding of the Hoogesten base pairing.
The presence of the carboxy moiety does not significantly alter the pKa of the N3 on cytidine with a pKa of 4.4 for cytidine versus a pKa of 4.0 for 5-carboxy-dC (Table 1), as determined by spectral analysis.6 The Watson-Crick hybridization of G-CCOO- is essentially unaffected by the modification.
| Nucleoside | pKa |
|---|---|
| dC | 4.4 |
| 5-methyl-dC | 4.5 |
| 5-carboxy-dC | 4.0 |
| 5-formyl-dC | 2.4 |
Incorporation of 5-carboxy-2’-deoxyCytidine into oligonucleotides is simple and does not require any changes to the coupling conditions. Deprotection requires the use of sodium hydroxide in methanol to prevent amide formation during the deprotection of the oligo.
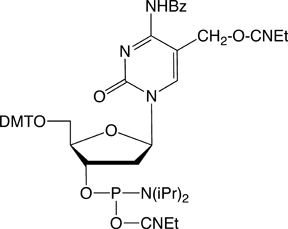
The duplex stability of oligos containing 5-formyl-dC is comparable to oligos containing 5-Me-dC, with the Tm for 5-Me-dC being approximately 1.3° C higher per incorporation.7 Interestingly, the misincorporation of thymidine opposite 5-formyl-dC was 3-4 times higher compared to control oligos containing dC and 5-Me-dC.7 The higher misincorporation rate was speculated to be a result of the changes in shape and the presence of the electron-withdrawing formyl group of 5-formyl-dC in comparison to 5-Me-dC. The overall effect is that 5-formyl-dC is considered highly mutagenic and increases the rate of transition mutations when evaluated by polymerase extension.7
In contrast to 5-carboxy-dC, the presence of the electron-withdrawing formyl group lowers the pKa to 2.4 compared to a pKa of 4.4 for dC and 4.5 for 5-Me-dC (Table 1).The hybridization of G-dCFo is unaffected by the modification as indicated by thermal denaturation.
The incorporation of 5-formyl-dC requires a 3 minute coupling time with 1H-tetrazole. Standard deprotection removes the 1,2-acetoxy protecting groups and subsequent oxidation with sodium periodate converts the 1,2-diol to formyl-dC, as shown in Figure 2.

5-Me-dC-CE Phosphoramidite (10-1060)
5-Hydroxymethyl-dC-CE Phosphoramidite (10-1062)
5-Carboxy-dC-CE Phosphoramidite (10-1066)
5-Formyl-dC-CE Phosphoramidite (10-1514)