When Mark Matteucci’s group at Gilead first described the cytosine analog AP-dC (Figure 1), they called it “G-clamp” because of its phenomenally high and specific affinity for guanosine. The G-clamp base modification recognizes both the Watson–Crick and Hoogsteen faces of the complementary guanine within a helix, leading to a large increase in duplex stability (Figure 2).1 Binding studies demonstrated that a single G-clamp substitution within an antisense oligonucleotide (ON) dramatically enhanced helical thermal stability and mismatch discrimination. Oligonucleotides containing G-clamp have been evaluated for sequence-context dependence, activity mismatch, sensitivity, RNAse-H cleavage, and hybridization kinetics in antisense experiments.
While Matteucci's aim was to improve antisense activity,2 there are two other properties of this remarkable cytidine analog that have not been widely examined. First, our work demonstrates that the AP-dC can successfully be used in primers. Second, the AP-dC has been observed to be a highly fluorescent nucleoside analog.



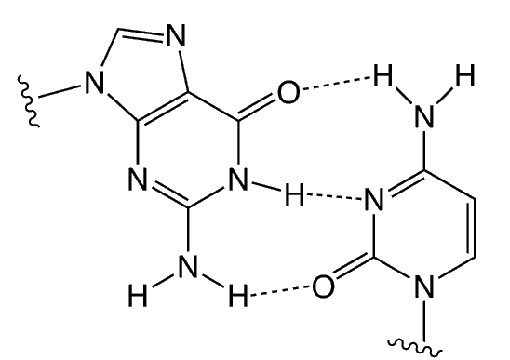

While much work has been done with AP-dC, the behavior of AP-dC-modified oligos in PCR amplification experiments was unknown. For this report, we examined the ability of oligonucleotides containing AP-dC to function as PCR primers and determined if Taq polymerase would incorporate dG opposite AP-dC in the template.
For this purpose, we synthesized several chimeric oligonucleotides that contained two parts: a universal sequence of 15 bases followed by a specific sequence of 20 bases targeting 3 S. cerevisiae ORFs (OAR1/YKL055c). The universal sequence contained standard nucleotides and the specific sequence included some AP-dC bases. The universal tags can then be used for sequencing the amplified fragments. The sequences are displayed in Table 1; positions of AP-dC are marked C.
| Primer name | Sequence |
|---|---|
| YKL055c.0.L | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.Lm1 | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.Lm2 | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.Lm3 | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.Lm4 | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.Lm5 | CGACGCCCGCTGATA GAAAGTATAGAGCGCACCGC |
| YKL055c.0.R | GTCCGGGAGCCATG CCAATGGTTCTCTCCAGCAT |
| YKL055c.0.Rm1 | GTCCGGGAGCCATG CCAATGGTTCTCTCCAGCAT |
| YKL055c.0.Rm2 | GTCCGGGAGCCATG CCAATGGTTCTCTCCAGCAT |
| YKL055c.0.Rm3 | GTCCGGGAGCCATG CCAATGGTTCTCTCCAGCAT |
| YKL055c.0.Rm4 | GTCCGGGAGCCATG CCAATGGTTCTCTCCAGCAT |
| ? | ? |
| primer Tag1 | GTCCGGGAGCCATG |
| primer Tag2 | CGACGCCCGCTGATA |
| ? | ? |
| specific S.cer. | GAA AGT ATA GAG CGC ACC GC |
| specific S.cer. | CCA ATG GTT CTC TCC AGC AT |
These primers were used to amplify S. cerevisae genomic DNA using standard Taq DNA polymerase.
The results displayed in Figure 3 correspond to an annealing temperature of 60°C and a magnesium concentration of 2 mM and 35 cycles. The lane named R-L corresponds to the PCR performed with control primers with no modification and is used as a positive control. R and L correspond to the non-modified primers. LmX and RmX indicate the use of various primers with one or more modifications (see Table 1 for sequence and modification positions.) Briefly, we see that when using a pair of primers containing AP-dC in one of the two primers, (R-LmX or L-RmX), we have amplification comparable to the control with no AP-dC except when using the most heavily modified primers in R-Lm4 (no amplification) and R-Lm5 (low amplification).
When modifications are present in both primers, we see a good amplification in primers containing the lowest number of total modifications (Figure 3 – lower panel). The results presented in Figure 3 are representative of many other combinations.
Based on these results we would conclude that it would be safe to limit:
However, we did not test enough situations to rule out that more AP-dC modifications could result in amplification with other thermostable polymerases or in other particular conditions.
Finally the results of PCR with Lm1 primer indicated that modification of the 3’-position with AP-dC is permitted for DNA amplification (R-Lm1 or Rm1-Lm1).
Several of the amplicons were extracted and sequenced using sequencing primers complementary to the universal tag using an ABI sequencer and Big Dye chemistry. The sequence alignments (data not shown) indicated that it was possible to obtain full length sequences covering the entire primer region and that no base modification was detected at positions opposite to any of the AP-dC. In each sequencing, we observed a dG opposite the AP-dC. Also, no particular sequencing problems were observed and the peaks corresponding to AP-dC positions had similar intensity compared to the control with dC at the same positions.
Substitution of AP-dC for dC can raise the Tm of shorter PCR primers or probes enough to amplify or probe in a region where it is normally difficult to obtain stable sequences due to length restrictions. We also look forward to its use in the development of short, easily prepared oligonucleotides for use in highly specific single nucleotide polymorphism (SNP) and other in vitro diagnostic assays.
With an extinction coefficient of approximately 10,500 M-1 and a quantum yield of fluorescence of 0.2, AP-dC is 2-3 times as bright as our popular Pyrrolo-dC analog. In addition, AP-dC exhibits a Stokes’ shift greater than 100 nm. As with most fluorescent base analogs, it is substantially quenched upon forming a duplex. The quantum yield drops to 0.1 while gaining significant structure in the emission spectrum (Figure 4), making it an ideal probe of DNA structure.


Glen Research thanks the oligo synthesis and DNA-array teams of Eurogentec (Seraing, Belgium) for making the oligos used in this study and for performing the PCR amplification and the DNA sequence analysis