Pyrrolo-dC is a fluorescent deoxycytidine analog that is an ideal probe of DNA structure and dynamics.

The fluorescence of a nucleoside base is highly dependent on the environment of the base and the measurement of its fluorescence is a powerful and sensitive tool for the analysis of DNA and RNA structure. It is especially useful for analyzing the interaction of DNA and RNA with their corresponding binding proteins. Fluorescence measurements allow real-time probing of these structural interactions. Unfortunately, the intrinsic fluorescence of the regular bases is extremely low to non-existent, so fluorescent analogues have been sought for a long time.
For probing DNA structure, the ideal fluorescent nucleoside:
Several fluorescent nucleoside analogues have been prepared as phosphoramidites in recent years. Etheno-A (1) and etheno-C (2)1 (Figure 1) are two readily accessible fluorescent structures but these molecules are both non-hybridizing. Other notable fluorescent base analogues are the pteridine nucleoside analogues actively being investigated by Pfleiderer, Hawkins and co-workers. The most promising analogue described to date2 is the adenosine analogue (3) but guanosine and other analogues have also been investigated.3-5


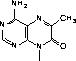
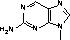
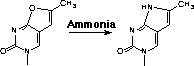
In 2001, Glen Research introduced furano-T (5) as a novel fluorescent nucleoside. It quickly became apparent that furano-T is unstable during cleavage and deprotection steps, but forms another fluorescent nucleoside on treatment with ammonium hydroxide. Mass spec data supported the conclusion that furano-T had been transformed by ammonium hydroxide in the cleavage and deprotection to the equivalent amino compound, pyrrolo-C (6) (Figure 1). Furano-dT-CE Phosphoramidite (7) (Figure 2) was discontinued and Pyrrolo-dC-CE Phosphor-amidite (8) (Figure 2), the fluorescent dC analogue, was introduced early in 2002. Pyrrolo-C CE Phosphoramidite (9) (Figure 2) for RNA synthesis has now also been synthesized and we foresee potential applications in RNA structural analysis.
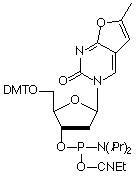
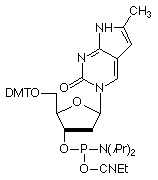
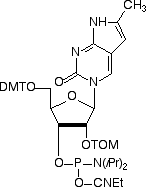
Pyrrolo-dC is stable to all DNA synthesis cycles and reagents, with the exception of strong iodine oxidizer solutions. Routinely, 0.02M iodine oxidizer solutions are now used and these solutions have no effect on pyrrolo-dC. However, some older instruments and cycles use 0.1M iodine or stronger solutions and these cause degradation of pyrrolo-dC each cycle. These strong iodine solutions should not be used with pyrrolo-dC.
Pyrrolo-dC is stable to most cleavage and deprotection conditions, including UltraMild with potassium carbonate in methanol or ammonium hydroxide at room temperature, and regular with ammonium hydroxide, preferably at room temperature. This analogue contrasts with the pyrrolo-dC derivative, lacking only the methyl group, described7 by Epoch researchers, which was unstable to deprotection conditions. They also observed that the non-methylated version of the ring system formed a mismatch with G, which differs from our observations for the pyrrolo-C - G base pair.
The spectral properties of pyrrolo-dC, coupled with its unique base-pairing ability, make this fluorescent analog extremely valuable in probing DNA structure. When the pyrrolo-dC is base-paired, its fluorescence is significantly quenched through what is most likely base stacking or dG interactions.
| QY | λ | e (L/mol·cm) | |
|---|---|---|---|
| single-stranded | 0.07 | 260 nm | 4000 |
| 347 nm | 3700 | ||
| double-stranded | 0.02 | ||
| (QY determined relative to quinine sulfate in 0.5M H2SO4) | |||
The quantum yield of fluorescence for pyrrolo-dC is quite sensitive to its hybridization state, making it ideally suited8,9 for probing the dynamic structure of DNA. Work by Liu and Martin has shown9 that, when the pyrrolo-dC is mismatched in an otherwise duplex hybrid, the fluorescence is higher than the single-stranded species when the mismatched base is adenosine. This most likely arises from efficient energy transfer from the adenosine to the pyrrolo-dC. This unusual behavior also allows differentiation in situ between a DNA-DNA duplex and a DNA-RNA heteroduplex.
The quenching of pyrrolo-dC allows local structural changes to be probed with great sensitivity. Using pyrrolo-dC, Liu and Martin8 have characterized the transcription bubble in elongation complexes of T7 RNA Polymerase to single-base resolution by observing roughly a two-fold increase in fluorescence as the polymerase induces melting. By starving the T7 RNA Polymerase of specific nucleoside triphosphates, the enzyme could be stalled at specific sites, producing 'fluorescence snapshots' of the complex, and yielding detailed information on the nature of the transcription bubble and heteroduplex.
Work is still progressing in evaluating the effect of this modified fluorescent nucleoside in biological systems and will be reported in detail later. However, a few comments on our findings to date may be of interest. Oligonucleotides containing pyrrolo-dC act as efficient primers and the PCR products appear to be identical for primers with 0 to 5 pyrrolo-dC residues replacing dC. Preliminary data indicate that pyrrolo-dC codes as dC in PCR experiments. And very preliminary evidence indicates that pyrrolo-dC triphosphate is incorporated efficiently by Taq polymerase and is incorporated specifically opposite dG.
 |
|
G ..... C Base Pair |
G ..... pyrrolo-C Base Pair |
We are happy to introduce pyrrolo-dCTP and look forward to its use in biological assay development.
The pyrrolo-dC project is a joint development by Berry and Associates (http://www.berryassoc.com) and Glen Research. Patents covering this modified fluorescent base and its uses are currently pending.
Pyrrolo-dC-CE Phosphoramidite (10-1017)
Pyrrolo-C-TOM-CE Phosphoramidite (10-3017)
Pyrrolo-dCTP (10mM) (81-1017) has been discontinued.