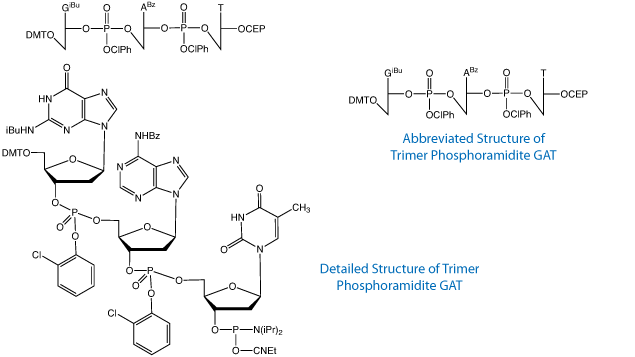
Trimer phosphoramidites, depicted in Figure 1, have proven to be extremely valuable because they allow codon-based mutagenesis, which circumvents the common problems of codon-bias, frame-shift mutations, and the introduction of nonsense or stop codons.1 This leads to the production of clonal libraries of exceptional diversity with order-of-magnitude increases in amino acid sequence variance while either maintaining a uniform amino acid distribution2 or one that is biased toward a desired set of amino acids.3 However, difficulties arise when trying to introduce mutations in multiple distal regions of a gene simultaneously. The synthesis of long oligonucleotides is required, which inevitably leads to lower sequence fidelity due to deletion mutants, depurination events and, to a lesser extent, mutations arising from deamination of cytidine, for example.
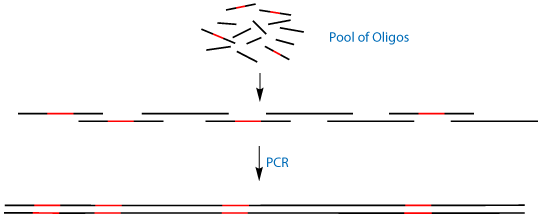
An elegant solution to this problem is the use of Antisense Trimer Phosphoramidites. These trimers are the reverse complement of the cannonical 'sense' codons. When these antisense codons are put into the noncoding strand of a template DNA and amplified by PCR, they will code for the sense codon in the opposite strand of DNA. This allows the powerful technique of PCR Assembly4 to generate not only kilobase-sized genes from short 50mer oligonucleotides, but to simultaneously mutate multiple distal regions of that gene, as shown in Figure 2.
The sense and their corresponding antisense codons are listed in Table 1. Conveniently, many of our existing sense trimers can act as antisense codons. For example, AAC, which codes for asparagine, has the anticodon GTT, which is the sense codon for valine. However, some of the existing trimers, while they can act as an antisense codon, are not good choices for use. For example, TGG, which codes for tryptophan, could be used as an antisense codon for proline because CCA is one of proline's synonymous codons. However, CCA has a relatively low Codon Adaptation Index (CAI) value5 in E. coli, which could limit protein expression in that commonly used organism. For this reason, the anticodon CGG was chosen for optimal expression in E. coli, as were the other new antisense codons shown in color in Table 1.
| Sense codons (5'->3') | Antisense codons (5'->3') | MW | RF (Temp) |
|---|---|---|---|
| AAA (Lys) | TTT | 1572.4 | 1.425 |
| AAC (Asn) | GTT | ||
| ACT (Thr) | GGT | ||
| ATC (Ile) | GAT | 1780.5 | 1.425 |
| ATG (Met) | CAT | ||
| CAG (Gln) | CTG | ||
| CAT (His) | ATG | ||
| CCG (Pro) | CGG | 1851.5 | 1.425 |
| CGT (Arg) | GCG | 1851.5 | 1.425 |
| CTG (Leu) | CAG | ||
| GAA (Glu) | TTC | ||
| GAC (Asp) | ATC | ||
| GCT (Ala) | TGC | ||
| GGT (Gly) | ACC | 1863.5 | 1.425 |
| GTT (Val) | AAC | ||
| TAC (Tyr) | GTA | 1780.5 | 1.425 |
| TCT (Ser) | AGA | 1893.5 | 1.425 |
| TGC (Cys) | GCA | 1869.5 | 1.425 |
| TGG (Trp) | CCA | 1863.5 | 1.425 |
| TTC (Phe) | GAA |
As with our existing trimers, the antisense trimer phosphoramidites are compatible with standard DNA synthesis reagents noting, however, that a special diluent is required, ACN/DCM 1:3 (v/v), to dissolve the phosphoramidite and a 15 minute coupling time is recommended. To prevent strand scission, oligos containing the antisense trimers should be deprotected in 30% ammonium hydroxide at room temperature for 17 hours, followed by an additional 4 hours at 55 °C to complete the deprotection of the nucleobases.
At present, we do not have enough data to accurately assign a Reaction Factor (RF) to the antisense trimers. Until those have been determined, a value of 1.425 is being used which is the average RF of the well-characterized sense trimers. As such, an antisense trimer mix designed to provide 20 amino acids represented equally would yield some trimers that were over represented (i.e., >5%) and others under represented (<5%) upon sequencing. We will update the RF values when the data become available.