Application note prepared by Richard T. Pon
June 11, 2007
The first nucleoside residue of an oligonucleotide sequence is always attached to the solid-phase support through a dicarboxylic acid linker arm. Traditionally, this linker has been succinic acid. However, the need for faster oligonucleotide processing led to the introduction of hydroquinone-O,O’-diacetic acid (“Q-Linker”) as a superior replacement. With the Q-Linker, all the steps required for synthesis and deprotection remain unchanged except for the time required to release the oligonucleotide product from the support. With oligodeoxyribonucleotides, the time required for the ammonium hydroxide cleavage step was reduced from 60 minutes to only 2 minutes, thus eliminating the need for multiple Nh4OH treatment steps and significantly increasing sample throughput. With ultra-fast deprotection using AMA (1:1 ammonium hydroxide and aqueous methylamine) the cleavage time is reduced to only a few seconds. In this case, the throughput gained using the Q-Linker is not as beneficial since AMA also cleaves the traditional succinyl linkers much faster (~ 5 minutes). However, we have found that this is not the case with RNA synthesis and, as described below, the Q-Linker provides significant benefits to RNA syntheses, even when using ultra-fast deprotection.
Recent discoveries of novel properties for short RNA sequences, such as siRNAs and miRNAs, have greatly increased the demand for chemically synthesized oligoribonucleotides. While the synthesis of oligoribonucleotides is very similar to oligodeoxyribonucleotides, the deprotection and work-up conditions are quite different. In particular, modified cleavage and base deprotection conditions are required to minimize loss of 2’-O-protecting groups and the subsequent chain cleavage which occurs; and to improve the solubility of the partially protected intermediates. The 2’-O-protecting group also slows the cleavage from the support through steric hindrance.
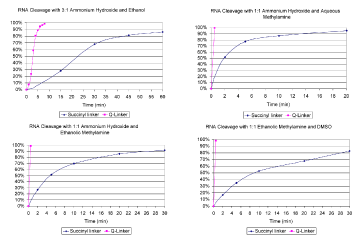
Therefore, reagents containing organic modifiers, such as ethanol or dimethyl sulfoxide (DMSO), are required. These eliminate chain cleavage and improved the solubility of long RNA products. However, these organic modifiers further decrease the rate of linker hydrolysis and cleavage times of 30-60 minutes at room temperature are required to recover most of the oligoribonucleotide product. Thus, cleavage of an RNA sequence is more than 15 times longer than cleavage of a DNA sequence, when ethanolic methylamine is used instead of aqueous methylamine in the AMA reagent. If the cleavage step is combined with the base deprotection step at elevated temperatures, then the products become contaminated with dissolved silica and separation of the partially deprotected oligoribonucleotide from the heterogeneous mixture containing the residual support is more time consuming and loss of product may also occur.
However, ultra-fast cleavage times can be restored by using the Q-Linker, as shown in Figures 1-4. In particular, ethanolic methylamine reagents only require cleavage times of less than 2 minutes for complete product recovery. The cleavage rate for our preferred RNA deprotection reagent (AMA with ethanolic methylamine) for both the succinyl and Q-Linker is shown below. Such fast cleavage at room temperature enables simplified sample processing (no multiple treatment and wait steps), greater sample throughput, reduced handling of volatile and noxious reagents, reduced risk of incomplete base deprotection (through accidental evaporation of methylamine or ammonia), and greater product recovery and quality. Most importantly, no other changes to the synthesis or deprotection procedures are required. The Q-Linker is compatible with all of the remaining steps in common RNA synthesis protocols.
Glen Research offers a variety of supports containing the Q-Linker under license from University Technologies International. We are also planning to add Q-supports optimized for RNA synthesis to our products in the near future.

For chemistry inquiries:
Professor Richard T. Pon
Dept. Biochemistry & Molecular Biology
University of Calgary
t: 403-220-4225
[email protected]